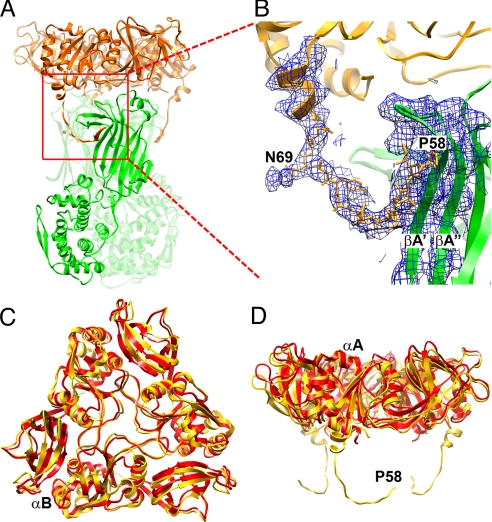
Fig. 2.
VP7 outer protein layer. (A) Structure of a VP6–VP7 heterohexamer, derived from crystal structures of VP6 (green) (14) and VP7 (gold) (16), docked into the cryo-EM density, and from the model built into cryo-EM density for the N-terminal arm. Two of the three VP7 N termini forming tight interactions with VP6 can be seen on the left and right side of the VP6 trimer. The red square indicates the area shown in more detail in B. (B) VP7 N terminus (residues 58–78) and corresponding cryo-EM density, showing its interaction with VP6. Pro-58 and Asn-69 are labeled; the latter bears a glycan for which density is present. Strands A′ and A″ of VP6 are also labeled. The density corresponding to residues 61–68 is weaker than in other parts of the map because this part of the structure lacks a clearly defined secondary structure and makes no contact with other parts of the structure. The map was filtered at 4.5-Å resolution to display a continuous density trace in this region. (C and D) Conformational differences between bound VP7 (gold) and the VP7 crystal structure (red) (16). The VP7 trimer is viewed from the outside of the virus along its symmetry axis (C) and from the “side”, normal to its symmetry axis (D). The way in which the domain hinge displacement “flattens” the subunit when the trimer binds VP6 is evident in the latter view. The alignment is based on a superposition of the Rossmann-fold domains.