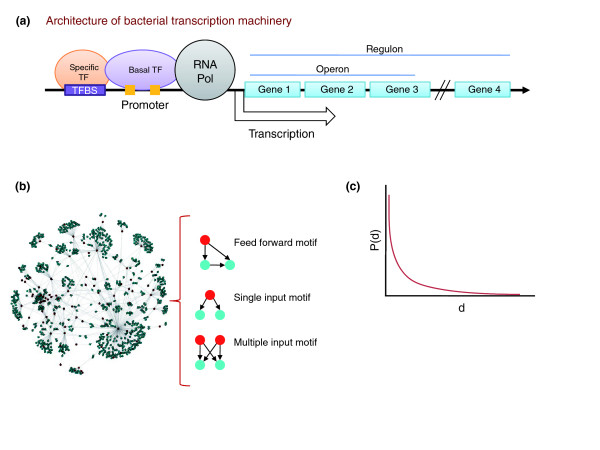
Figure 1.
The transcription apparatus and transcription regulatory network of bacteria. (a) Schematic representation of the architecture of bacterial transcription machinery and operons and regulons. A regulon is the set of genes regulated by one transcription factor; an operon is a set of adjacent genes transcribed into one mRNA. (b) Architecture of transcription regulatory networks. The global structure (left) and three types of motifs found in transcription regulatory networks (right) are depicted as ordered graphs. Red dots indicate transcription factors; blue dots indicate targets. (c) The degree distribution of transcription factor-target interactions is approximated by a power-law equation [5]. The graph shows a power-law distribution; degree (d) is the number of regulatory connections between a transcription factor and target genes, while P(d) indicates the probability of transcription factors with a particular number of such connections. Pol, polymerase; TF, transcription factor; TFBS, transcription factor binding site.