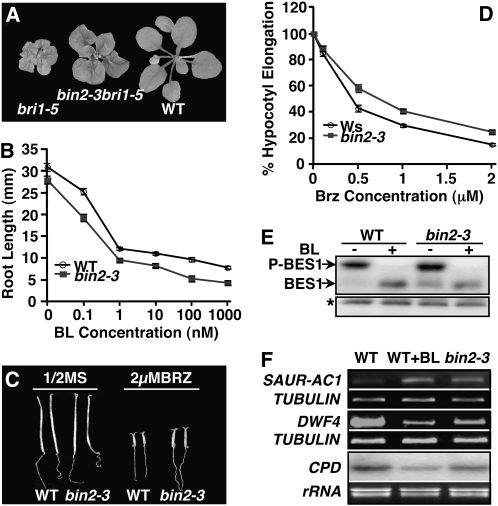
Figure 2.
Loss of BIN2 function stimulates BR signaling. A, The T-DNA insertional bin2-3 mutation partially suppresses the bri1-5 mutation. Shown from left to right are 4-week-old soil-grown plants of bri1-5, bin2-3bri1-5, and the Ws wild-type (WT) control. B, Root lengths of 2-week-old seedlings of the wild type and bin2-3 grown on half-strength MS medium containing no or increasing concentrations of BL. C, Six-day-old etiolated seedlings of the Ws wild type and bin2-3 grown on medium containing no or 2 μm Brz. D, Hypocotyl elongation of wild-type and bin2-3 seedlings grown on medium containing different concentrations of Brz relative to seedlings of the same genotype grown on regular medium (see “Materials and Methods” for details). For both B and D, each data point represents an average of 60 seedlings of two duplicate experiments. Error bars represent se. E, Western blot analysis of BES1 phosphorylation status. Eighteen-day-old seedlings were treated with 1 μm BL or mock solution for 1 h. Total proteins were extracted and analyzed by western blotting with anti-BES1 antibody. BES1-P is the phosphorylated BES1, and the asterisk indicates a nonspecific band for a loading control. F, RT-PCR and northern-blot analyses of SAUR-AC1, DWF4, and CPD gene expression in 18-d-old seedlings treated with or without 1 μm BL. See “Materials and Methods” for experimental details. β-TUBULIN was used as a control for RT-PCR analysis of both SAUR-AC1 and DWF4, while ethidium bromide staining of rRNAs served as a loading control for northern-blot analysis of CPD.