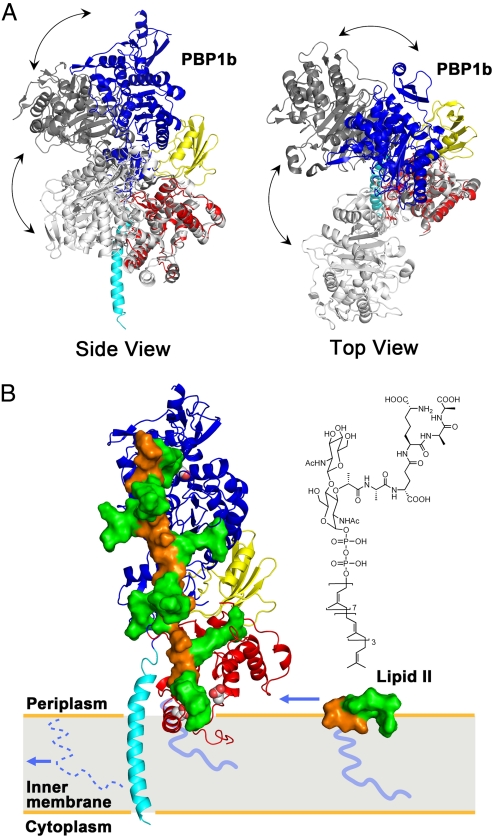
Fig. 4.
Interdomain flexibility and a model of peptidoglycan synthesis. (A) E. coli PBP1b and 2 SaPBP2 conformers (6, 23) were indicated by colored, light gray (PDB ID code 3DWK), and dark gray (PDB ID code 2OLV), respectively. The TG domain of SaPBP2 was superimposed onto the TG of E. coli PBP1b. Side view (Left) and top view (Right) of the comparison reveals possible flexibility of a hinge region between TG and TP domains. (B) Active sites of TG and TP were shown by van der Waals spheres. The disaccharide, pentapeptide, and lipid tail of lipid II and peptidoglycan were shown in orange surface, green surface, and blue line, respectively. A single strand of the proposed peptidoglycan model (25) was docked onto the structure of E. coli PBP1b, with lipid IV portion replacing moenomycin. The incoming lipid II, of which the chemical structure is shown on top, diffuses in the plane of the membrane. After the TG reaction, the lipid moiety of this lipid II is kept as the membrane anchor, whereas the original lipid tail (shown as dotted line) is recycled. The polymerized peptidoglycan grows perpendicularly to the membrane and toward the TP domain, where the cross-linking reaction of the pentapeptides takes place (see also Movie S3).