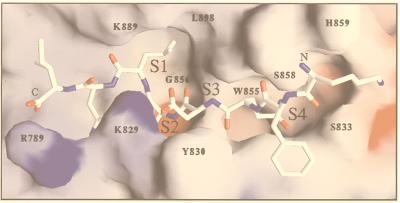
Figure 8.
Homology model of PfSUB2 predicted catalytic domain in complex with MSP1–42 cleavage site (Nterm-K-F-Q-D-M-L-N-I-Cterm). Residues involved in the PfSUB2 predicted S1, S2, S3, and S4 subsites are numbered. Modeling was carried out with computer programs quanta and charmm by using the atomic coordinates of B. amyloliquefaciens BPN subtilisin in complex with the eglin inhibitor (PDB code 2SNI) and B. licheniformis Carlsberg’s subtilisin in complex with eglin C (PDB code 2SEC) as templates. Framework residues differing between proteins were mutated, and all required insertions (Fig. 1) were modeled in hypothetical conformations taken from the loop library implemented in quanta. These insertions occur in surface-exposed loops located far from the substrate-binding region, with the possible exception of the segment preceding the catalytic His793 residue (including Arg789), which may influence the local structure of the S1′-S2′ subsites. The model was manually adjusted to remove unreasonable contacts and subjected to energy minimization by using the program charmm.