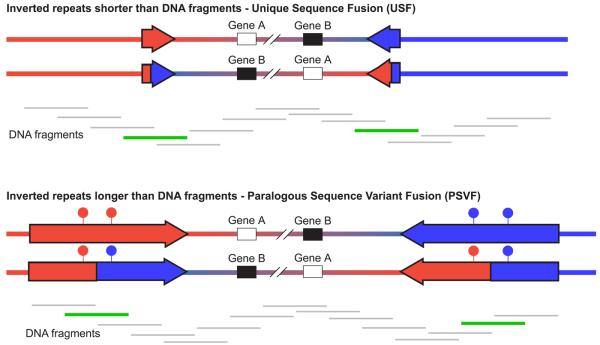
Figure 1. Strategies for genotyping inversions.
Inverted and wildtype orientations are represented for two inversions. In both cases the inversion breakpoints lie within inverted repeats, shown as red and blue arrows. In the first case, the inverted repeats are shorter than the size of the DNA fragments in the genomic DNA preparation, whereas in the second case the inverted repeats are longer than the DNA fragments. Beneath each inversion are shown the DNA fragments that cover the inverted region. DNA fragments that are informative for genotyping the inversion are shown in green, whereas those that are uninformative are shown in grey. The filled circles above the inverted repeats in the second inversion represent paralogous sequence variants that distinguish between the two repeats; the haplotype of which is diagnostic for the state of the inversion.