Abstract
Prodrug activation gene therapy is a developing approach to cancer treatment, whereby prodrug-activating enzymes are expressed in tumour cells. After administration of a non-toxic prodrug, its conversion to cytotoxic metabolites directly kills tumour cells expressing the activating enzyme, whereas the local spread of activated metabolites can kill nearby cells lacking the enzyme (bystander cell killing). One promising combination that has entered clinical trials uses the nitroreductase NfsB from Escherichia coli to activate the prodrug, CB1954, to a potent bifunctional alkylating agent. NfsA, the major E. coli nitroreductase, has greater activity with nitrofuran antibiotics, but it has not been compared in the past with NfsB for the activation of CB1954. We show superior in vitro kinetics of CB1954 activation by NfsA using the NADPH cofactor, and show that the expression of NfsA in bacterial or human cells results in a 3.5- to 8-fold greater sensitivity to CB1954, relative to NfsB. Although NfsB reduces either the 2-NO2 or 4-NO2 positions of CB1954 in an equimolar ratio, we show that NfsA preferentially reduces the 2-NO2 group, which leads to a greater bystander effect with cells expressing NfsA than with NfsB. NfsA is also more effective than NfsB for cell sensitisation to nitrofurans and to a selection of alternative, dinitrobenzamide mustard (DNBM) prodrugs.
Gene-directed enzyme prodrug therapy (GDEPT) is a developing strategy for cancer treatment, involving delivery to tumour cells of an exogenous gene, encoding an enzyme that can convert a non-toxic prodrug into cytotoxic products. In principle, local generation of highly reactive cytotoxins within the cancer cells allows optimal therapeutic effect, whereas systemic toxicity remains lower than with conventional chemotherapy (McNeish et al, 1997; Niculescu-Duvaz and Springer, 2005; Russell and Khatri, 2006). As specific gene delivery to all tumour cells seems unattainable using current methods, the ability of an activated prodrug to spread locally from cell to cell is considered an essential feature for a successful GDEPT system. It allows tumour cells that have escaped gene transfer to be killed as a result of prodrug activation in nearby cells that express the activating enzyme. This is known as the ‘bystander effect’ (Freeman et al, 1993).
CB1954 (5-[aziridin-1-yl]-2,4-dinitrobenzamide) is a prodrug, which is converted from a weak, monofunctional alkylating agent to a potent bifunctional alkylating agent upon nitroreduction (Knox et al, 1988, 1991; Friedlos et al, 1992). Anlezark et al (1992) reported that the nitroreductase encoded by the nfsB gene of Escherichia coli could activate CB1954, leading to the initial adoption of this enzyme for use with CB1954 in GDEPT (Anlezark et al, 1992; Bridgewater et al, 1995; Grove et al, 1999; Searle et al, 2004). The expression of NfsB in cancer cells using replication-defective retrovirus or adenovirus vectors was shown to confer greatly increased sensitivity to CB1954, and anti-tumour activity was shown in vivo (McNeish et al, 1998; Djeha et al, 2000, 2001; Weedon et al, 2000). After the initial phase I clinical trials of CB1954 alone (Chung-Faye et al, 2001), and of a replication-defective adenovirus expressing NfsB (CTL102) (Palmer et al, 2004), the combination has been tested in a phase I/II trial in patients with prostate cancer. A decline in serum levels of prostate specific antigen in some patients suggests anti-tumour activity (Patel et al, 2009); however, greater efficacy is desirable.
NfsB is a homodimeric flavoenzyme which can use either NADH or NADPH to reduce the tightly bound FMN; after dissociation of the NAD(P)+; a variety of nitroaromatic or quinone substrates can be reduced in a second reaction step (Anlezark et al, 1992; Lovering et al, 2001; Race et al, 2005). The published Km for NfsB nitroreductase with CB1954 is 862 μM (Anlezark et al, 1992), which is considerably higher than its peak serum concentration achievable in humans (5–10 μM) (Chung-Faye et al, 2001). Thus, the activation of CB1954 by NfsB in vivo will be very inefficient, justifying the consideration of alternative enzymes (Anlezark et al, 2002). NfsB was originally identified through its role in bacterial sensitivity to nitrofuran antibiotics (Sastry and Jayaraman, 1984). Selection of E. coli for nitrofurazone resistance leads first to mutations in the major nitroreductase gene, nfsA (Whiteway et al, 1998), and only subsequently in nfsB. Alignment of the amino acid sequences shows only 28 identical and 32 conserved residues out of the 242 or 217 residues of NfsA and NfsB, respectively, and antibodies specific for NfsB do not cross-react with NfsA (unpublished results). The two enzymes share many structural features, including a 5-stranded anti-parallel β-sheet core and surrounding α-helices, with the two active sites occupying clefts at the dimer interface and presenting the re-face of the FMN isoalloxazine ring towards the substrate pocket (Kobori et al, 2001; Lovering et al, 2001). Although some residues around the active site are conserved, the active site of NfsA is more open than that of NfsB, an observation that contributed to our decision to investigate its activity with CB1954 and other prodrugs. Kinetic studies of the purified enzymes showed that NfsA has a two- to three-fold greater activity with nitrofurazone and several other nitroaromatic substrates, compared with NfsB (Zenno et al, 1996). The report that NfsA has a marked preference for the cofactor NADPH (Zenno et al, 1996) suggested that this enzyme may have been overlooked in the initial isolation of NfsB for the reduction of CB1954, which used NADH as a cofactor (Anlezark et al, 1992). One study has shown that NfsA is capable of CB1954 activation (Barak et al, 2006); however, to our knowledge, no earlier study has compared the activities of NfsA and NfsB with CB1954. In this study, we compare the ability of these two E. coli nitroreductases to sensitise cells to CB1954 and a selection of other prodrugs. We also compare their kinetics of CB1954 activation in vitro, and show that NfsA preferentially reduces the 2-NO2 group of CB1954, resulting in an improved bystander cell killing. Overall, the results suggest that NfsA could have advantages over NfsB for use in GDEPT with CB1954 or several other nitroaromatic prodrugs.
Materials and methods
Bacterial expression vector construction
The nfsA gene was amplified by PCR from E. coli DH5α genomic DNA using primers PS1296C GGAATTCATATGACGCCAACCATTGAACTTATTTGTG and PS1296D GTGGATCCTATTAGCGCGTCGCCCAACCCTG, digested with Nde I and Bam HI (underlined) and inserted between these sites downstream of the tac promoter in plasmid pJG12B1 (Grove et al, 2003), producing pSV036B9. The same Nde I to Bam HI fragment was also inserted into pET24c (Novagen, Madison, WI, USA), making pPS1341A1. The Sfi I fragment containing the nfsA gene and the upstream ribosome binding site from pSV036B9 was ligated downstream of the tac promoter into the bacteriophage λ-vector λJG3J1 (Grove et al, 2003), producing λSV054, which was packaged and used to generate lysogens of the E. coli strain, UT5600 (nfsB−), as before (Grove et al, 2003).
Bacterial prodrug susceptibility assay
Cultures of E. coli UT5600 lysogens carrying λSV054, the empty vector λJG3J1 or the same vector expressing NfsB (λJG16C2) were grown to mid log phase in a liquid medium, before dilution and spreading onto agar plates containing a range of CB1954 concentrations, essentially as described (Grove et al, 2003). Colonies were counted after 24 h of incubation at 37°C. The plating efficiency at each prodrug concentration was calculated as a percentage of the number of colonies obtained in the absence of prodrug.
Protein purification
NfsB was expressed in the E. coli strain BL21(λDE3) transformed with a pET11c plasmid containing the nfsB gene and purified as described (Lovering et al, 2001). NfsA was purified from E. coli BL21(λDE3) transformed with plasmid pPS1341A1, using a method based on that used for NfsB, but the initial supernatant after sonication was fractionated using 30–70% NH4SO4. The 70% NH4SO4 precipitate was dissolved and fractionated on phenyl sepharose and Q-sepharose columns as for NfsB (Lovering et al, 2001).
Steady-state enzyme kinetic studies
Reactions were performed in 10 mM Tris pH 7.0, 5% DMSO at 25°C, initiated by the addition of a small volume of cold, dilute enzyme (typically 10 nM), and the initial rates of formation of the 2- and 4-hydroxylamine products of CB1954 were monitored spectroscopically at 420 nm, a wavelength at which both products have the same molar absorbance (1200 M−1cm−1). The data were fitted to Michaelis–Menten curves using SigmaPlot 10 (Systat Software Inc., Richmond, CA, USA) as described earlier (Race et al, 2007).
HPLC analysis of CB1954 reaction products
Reaction mixtures containing 200 μM CB1954, 1 mM NAD(P)H and 1 μM NfsA or NfsB in 10 mM Tris pH 7.0, 5% DMSO were incubated at room temperature for 5 min, before separation by semi-preparative reverse-phase HPLC as described earlier (Race et al, 2007). To confirm the identity of the products, their absorbance was monitored simultaneously at four different wavelengths (246, 260, 310 and 397 nm) each at or close to absorbance maxima for one or more product. The amount of 2- and 4-hydroxylamine products isolated using this system was determined from the peak areas of the chromatogram, using ε260 nm=7880 M−1cm−1 for the 4-hydroxylamine and ε260 nm=5420 M−1cm−1 for the 2-hydroxylamine (Knox et al, 1991).
For the analysis of cellular metabolites of CB1954, single cell suspensions of SKOV3-GFP, SKOV3-NfsB and SKOV3-NfsA stable cell lines were generated from log phase monolayer cultures and stirred magnetically at 5 × 105 cells per ml in an αMEM culture medium (Sigma-Aldrich Co. Ltd, Gillingham, UK) without serum and supplemented with additional ascorbate (50 μg ml−1) to prevent the auto-oxidation of the hydroxylamine metabolites. After equilibration, CB1954 was added to a final concentration of 10 or 100 μM. Samples (300 μl) were removed at various time points up to 3 h, centrifuged rapidly and the extracellular medium was stored at −80°C until analysed by HPLC using detection at 254 nm (peak identity confirmed by reference to authentic standards) or by LC/MS (4-amine metabolite, detected using negative mode atmospheric pressure chemical ionisation at m/z 221) as described (Helsby et al, 2004).
Sensitisation of SKOV3 cells to CB1954 using purified enzymes
SKOV3 human ovarian carcinoma cells were seeded at a density of 10 000 cells per well in 96-well plates, in Dulbecco's Modified Eagle's Medium (Sigma-Aldrich Co. Ltd) (HEPES buffered) with 10% foetal calf serum, and incubated at 37°C in 5% CO2. After 2 days, the medium was removed from the wells and replaced with 50 μl of serum-free medium containing 100 nM FMN and various concentrations of purified NfsA or NfsB, to which was added 100 μl of serum-free medium containing CB1954 and cofactor (NADH or NADPH), to give final concentrations of 0.1–100 nM enzyme, 50 μM CB1954 and 200 μM cofactor. The cultures were incubated for 4 h at 37°C before the prodrug-containing medium was replaced with fresh, complete medium. The relative number of viable cells was determined after a further 2 days using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Mosmann, 1983). Absorbances at 490nm were read on a Wallac Victor 2 1420 Multilabel Counter (Perkin-Elmer, Fremont, CA, USA), normalised relative to 100% survival of cells in wells with no added enzyme. Sigmoid curves were fitted, and F-tests performed to determine the statistical significance of differences between the curves, using Graphpad Prism software (GraphPad Software Inc., La Jolla, CA, USA).
Eukaryotic expression: retroviral vectors
The retrovirally transduced SKOV-NTR cell line, which stably expresses NfsB, was described earlier (McNeish et al, 1998). For clarity in this report, SKOV-NTR cells are referred to as SKOV3-NfsB.
The nfsA gene was amplified by PCR from E. coli DH5α genomic DNA using primers PS1296A GCCGCCACCATGACGCCAACCATTGAACTTATTTGTG and PS1296D, and inserted into the Hpa I site downstream of the CMV promoter in the retroviral vector plasmid pxLNCX (Green et al, 1997) to generate pSV035. The retrovirus packaging cell line, FLYA13, (Cosset et al, 1995) was transfected with pSV035 using calcium phosphate (Graham and van der Eb, 1973), and retrovirus producer cells were selected using G418 (500 μg ml−1). The supernatant from these cells was filtered (0.45 μm) and added with 8 μg ml−1 of polybrene (Sigma-Aldrich Co. Ltd) to SKOV3 ovarian carcinoma cells, which were subsequently selected using G418 (500 μg ml−1). Clonal cell lines were generated by limiting dilution; SKOV3-NfsA clone 4 was found to have a CB1954 sensitivity very close to that of SKOV3-NfsB, and was chosen for further experiments. As a negative control, a clone of SKOV3 cells (SKOV3-GFP) was generated with a similar vector expressing enhanced green fluorescent protein (EGFP).
Adenovirus vectors
The E1-, E3-deleted, replication-defective adenovirus CTL102, which expresses NfsB from the CMV promoter, was described earlier (Djeha et al, 2001). An equivalent adenovirus AdSV042 expressing NfsA was generated similarly, using the Bam HI fragment containing the CMV promoter and the nfsA gene from pSV035, inserted into the E1 deletion. Transfection, two rounds of plaque purification, and subsequent virus expansion used 911 cells (Fallaux et al, 1996). Virus was purified by CsCl banding, viral particle concentration was determined by DNA assay using Picogreen (Invitrogen, Paisley, UK), and infectious titre (p.f.u. ml−1) was determined using a plaque assay on 911 cells.
Cytotoxicity assays
SKOV3 cells were infected in suspension with the adenovirus vectors at multiplicities from 30 to 300 p.f.u. per cell for 90 min, before plating 1 × 104 cells per well in 96-well plates. After 2 days, the medium was replaced with fresh medium containing prodrug at a range of concentrations. Triplicate wells were used for each condition. The prodrug was removed after 4 h incubation at 37°C and replaced with fresh medium. Cell survival was determined using the MTT assay 48 h later (Mosmann, 1983). Absorbances were read on a Victor plate reader, normalised relative to 100% survival of cells not exposed to prodrug, and sigmoid curves were fitted, and the significance of differences between the curves was determined by the F-test, using the Graphpad Prism software.
To determine the bystander effect with NfsA vs NfsB, SKOV3-NfsA or SKOV3-NfsB cells were mixed with SKOV3 cells at proportions of 100, 50, 25, 12.5, 6.25, 3.13, 1.56 and 0%, and plated at a total of 2 × 104 cells per well in 96-well plates. After 1 day, the cell mixtures were exposed to a range of CB1954 concentrations in triplicate, for 18 h. Cell survival was assayed using the MTT assay 48 h later.
Comparison of the cytotoxicity of DNBM prodrugs with SKOV3-NfsA and SKOV3-NfsB cells used 6 × 103 cells per well plated the day before the addition of prodrug. The medium was replaced after 4 h of prodrug exposure, and the relative cell number was determined 4 days after the addition of prodrug by staining with sulphorhodamine B (Skehan et al, 1990).
Results
Sensitisation of E. coli to CB1954 by overexpression of NfsA nitroreductase
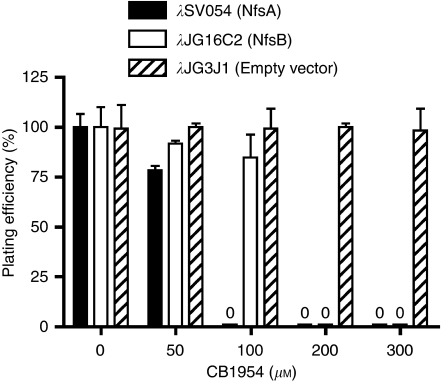
We inserted nfsA into a bacteriophage λ-vector under the control of the IPTG-inducible tac promoter and generated E. coli lysogens, in which a single copy of the bacteriophage genome was inserted into the bacterial chromosome. The expression of NfsB in this system sensitises the bacteria to CB1954, resulting in the inhibition of colony formation when the bacteria are plated on agar containing CB1954. The reduction in plating efficiency depends both on the concentration of CB1954 and on the activity of the expressed enzyme with the prodrug (Grove et al, 2003; Guise et al, 2007).
As shown in Figure 1, bacterial lysogens carrying the empty vector (λJG3J1) showed no reduction in plating efficiency at CB1954 concentrations up to 300 μM, indicating that the expression levels of endogenous NfsA or other enzymes do not confer significant sensitisation to the prodrug over this range. Lysogens carrying the vector λJG16C2, which expresses NfsB, showed complete inhibition of colony formation at 300 and 200 μM CB1954, but at 100 μM CB1954 and below the plating efficiency was ⩾80%. Bacteria carrying λSV054, expressing NfsA, showed greater sensitisation to CB1954, with complete inhibition of colony formation at ⩾100 μM CB1954, implying that NfsA catalyses the activation of CB1954 more efficiently than does NfsB.
Figure 1.
Sensitisation of Escherichia coli to CB1954 by the expression of NfsA or NfsB. Log phase cultures of E. coli UT5600 stably lysogenised with bacteriophage λ-vectors expressing NfsA or NfsB, or empty vector as control, were diluted and plated on agar containing 0, 50, 100, 200 or 300 μM CB1954. Colonies were counted after 24 h, and expressed as a percentage (%) of the number obtained from the same liquid culture plated in the absence of a prodrug (mean and range of duplicates).
In vitro kinetics of NfsA and NfsB with CB1954
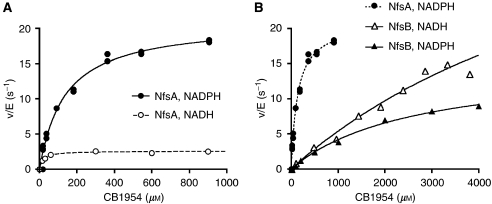
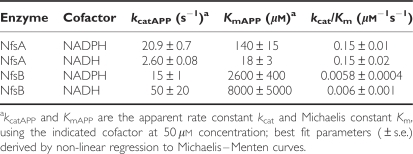
Figure 2 compares the rates of CB1954 activation by purified NfsA and NfsB as a function of CB1954 concentration, using either NADH or NADPH as a cofactor. In agreement with earlier work (Zenno et al, 1996), NfsA has a higher catalytic activity with NADPH than with NADH. At 50 μM cofactor, this results in ∼8-fold higher maximum rate of CB1954 activation with NADPH (Figure 2A). For NfsB, there is a small preference for NADH over NADPH, with a <2-fold difference in the rate of CB1954 activation for the two cofactors over the achievable solubility range of CB1954 (Figure 2B, solid lines). With its preferred cofactor NADPH, NfsA shows a much greater efficiency of CB1954 activation than NfsB, as shown by the dotted curve in Figure 2B. At the low substrate concentrations achievable in vivo (<10 μM), the rate of reaction is proportional to kcat/Km; for NfsA this is ∼25-fold higher than for NfsB (see Table 1 ).
Figure 2.
In vitro kinetics of CB1954 activation by purified NfsA and NfsB, using NADH or NADPH as cofactors. Graphs plot initial rate of CB1954 activation (v) normalised by enzyme concentration (E), using 50 μM NAD(P)H at a range of CB1954 concentrations, with Michaelis–Menten curves fitted to the data. (A) Kinetics of NfsA. (B) Kinetics of NfsB (solid lines); for comparison, the dotted line again shows the kinetics of NfsA using NADPH.
Table 1. Kinetic parameters for reduction of CB1954 by NfsA or NfsB, using 50 μM NADH or NADPH as a cofactor.
Owing to solubility limits of CB1954 in an aqueous solution, 5% DMSO was included in the reaction buffer to achieve CB1954 concentrations >1 mM. In separate experiments using 100 μM CB1954, 5% DMSO was found to slow the rate at which NfsA and NfsB reduced CB1954 by 10 and 35%, respectively. Thus, the data of Figure 2 and Table 1 may overestimate the kinetic advantage of NfsA by a factor of ∼1.4; nevertheless, this still implies that NfsA will activate CB1954 ∼18-fold faster than NfsB in physiological conditions.
Increased sensitisation of human carcinoma cells to CB1954 by NfsA nitroreductase
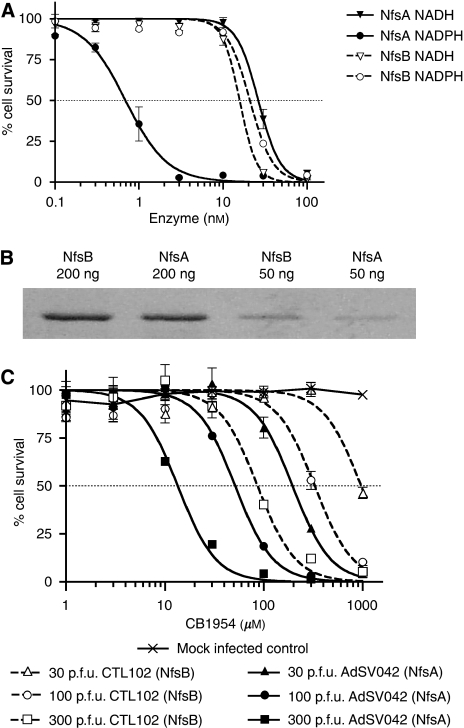
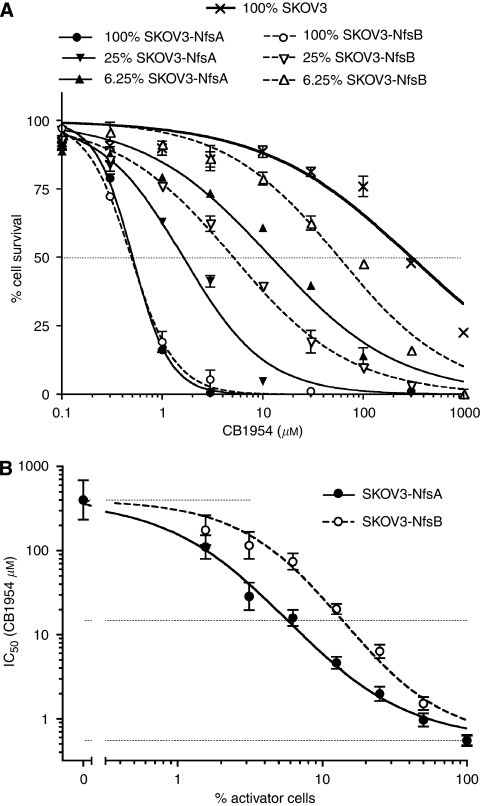
To compare the relative potency of NfsA and NfsB for sensitising human cancer cells with CB1954, we incubated SKOV3 ovarian carcinoma cells for 4 h with 50 μM CB1954 and either NADH or NADPH (200 μM), together with a range of concentrations of the purified enzymes. Cell viability was measured 2 days later. As shown in Figure 3A, at 100 nM, either of the enzymes with either cofactor could activate sufficient CB1954 to kill all the cells, but below 10 nM enzyme, only NfsA achieved significant toxicity, and that required NADPH as a cofactor. Sigmoid curves fitted to the data indicate that a 50% reduction in cell viability would require just 0.69 nM NfsA (95% CI: 0.62–0.77) when used with NADPH, whereas equivalent cell killing requires 15.7 nM NfsB (with NADH; 95% CI: 14.3–17.3) or 20.8 nM NfsB (with NADPH, 95% CI: 19.1–22.7). As indicated by the kinetic studies, NfsA was much less effective with the NADH cofactor, requiring 26.3 nM enzyme (95% CI: 24.9–27.7) to kill half the cells. The accuracy of the enzyme concentrations was confirmed by running 200 and 50 ng of each enzyme on a SDS-polyacrylamide gel, which was stained with Coomassie blue (Sigma-Aldrich Co. Ltd) (Figure 3B). Thus, these data confirm that NfsA could be up to 20-fold more effective than NfsB for prodrug activation gene therapy with CB1954, although clearly the actual benefit will depend on the relative availability of the reduced cofactors in the cell.
Figure 3.
Sensitisation of human ovarian carcinoma cells to CB1954 by NfsA or NfsB. (A) SKOV3 cells were incubated in a medium containing 50 μMCB1954, 200 μM NADH or NADPH and a range of concentrations of purified NfsA or NfsB proteins. The components were mixed at the start of a 4-h incubation period at 37°C, after which the medium was changed. Cell viability was determined 2 days later using the MTT assay. One representative experiment of three is shown. (B) To confirm the enzyme concentrations used in panel A, 200 and 50 ng of the purified proteins were analysed by SDS-PAGE and stained with Coomassie blue. (C) SKOV3 cells were infected using 0, 30, 100 or 300 p.f.u. per cell of adenovirus vectors CTL102 or AdSV042, expressing NfsB or NfsA, respectively. After 2 days, they were exposed to a range of CB1954 concentrations for 4 h, before determination of cell viability using MTT assay 2 days later. (The values used are representative of at least three experiments.)
To investigate cell sensitisation to CB1954 by intracellularly expressed enzymes, we constructed a replication-defective adenovirus (AdSV042) expressing NfsA nitroreductase from the CMV promoter, and compared this with the similar NfsB-expressing virus, CTL102 (Djeha et al, 2001). SKOV3 ovarian carcinoma cells were infected with 30, 100 or 300 p.f.u. per cell of the two viruses, or mock-infected, and after 2 days, the cultures were exposed to a range of CB1954 concentrations for 4 h. Dose-response curves showing the effect of CB1954 on cell viability 2 days later are shown in Figure 3C. Mock-infected cells showed no reduction in viability even with 1000 μM CB1954, whereas cells infected with the nitroreductase-expressing adenoviruses were sensitised to prodrug. As expected, greater sensitisation was achieved by higher multiplicity of infection (MOI) of the viruses. At each MOI, greater sensitisation was achieved by the virus expressing NfsA. For example, the IC50 (prodrug concentration required to reduce viable cell number to 50%) for cells infected with 300 p.f.u. per cell AdSV042 was 13.5 μM (95% CI: 13.0–14.0), whereas that obtained with 300 p.f.u. per cell CTL102 was 86.6 μM (95% CI: 82.9–90.6), corresponding to a 6.3-fold greater sensitisation by the virus expressing NfsA (P<0.0001). In other similar experiments, the benefit of expressing NfsA rather than NfsB by the virus at this MOI ranged from 3.5 to 8.1 fold. To determine the proportion of SKOV3 cells infected by adenovirus vectors at these MOI, we used a similar adenovirus expressing enhanced green fluorescent protein (Ad-EGFP), and analysed the infected cell population by flow cytometry (data not shown). Two days after infection with 300 p.f.u. per cell, 99% of the infected cell population expressed detectable EGFP, indicating that the prodrug-dependent cytotoxicity achieved with this MOI of the NfsA- or NfsB-expressing viruses can be attributed to the expression of the enzymes in almost every cell. Infection with 100 or 30 p.f.u. per cell Ad-EGFP resulted in 87 and 52% EGFP-positive cells, respectively. Thus, the ability of higher concentrations of CB1954 to kill more than 87 or 52% of the cells infected with 100 or 30 p.f.u. per cell of the nitroreductase-expressing viruses is indicative of bystander cytotoxicity, attributable to the escape of activated prodrug from nitroreductase-expressing cells.
Metabolite analysis of CB1954 after activation by NfsA nitroreductase
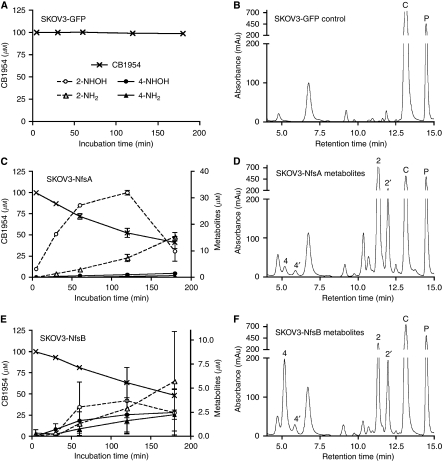
In the past, NfsB nitroreductase was shown to reduce either of the two nitro groups of CB1954 to the hydroxylamines, in equal proportions (Knox et al, 1992; Race et al, 2007); greater cytotoxicity was reported for the 4-NHOH derivative in DNA repair-deficient hamster cell lines (Knox et al, 1988, 1991; Helsby et al, 2003), although for a number of human cancer cell lines, both the 4-NHOH and 2-NHOH (and derived 2-NH2) products were reported to have similar cytotoxicity (Helsby et al, 2004). We used HPLC to compare the products of CB1954 reduction by NfsA and NfsB, both with purified enzymes and in cells. Supplementary Figure 1 shows the products of reduction of CB1954 by the purified enzymes. Purified NfsB generated equal amounts of the 2-NHOH and 4-NHOH products from CB1954, whereas NfsA only generated significant amounts of the 2-NHOH species; no other reduction products were observed.
To compare the prodrug activation products generated by the enzymes in the intracellular environment, the metabolites released to the medium by SKOV3 cells stably expressing NfsA or NfsB, or control cells (expressing GFP) were examined at different times after the addition of CB1954 (Figures 4A, C and E). Figures 4B, D and F show HPLC traces corresponding to the 2 h time point. The HPLC profiles show some peaks in addition to CB1954 (from media components) present in the supernatant of the control, GFP-expressing cells (Figure 4B). However, there was no reduction in the CB1954 peak during the incubation, and no metabolites of CB1954 were detected in the supernatant from SKOV3-GFP cells (Figure 4A). Two additional major peaks were produced in the medium from SKOV3-NfsA cells (Figure 4D), corresponding to the 2-NHOH and 2-NH2 derivatives of CB1954. As shown in the time course (Figure 4C), accumulation of the 2-NH2 species in supernatant of SKOV3-NfsA cells lags behind that of the 2-NHOH, as expected for a further reduction product from the hydroxylamine. As the amine metabolites of CB1954 are not detected after reduction by purified NfsA or NfsB, they may result from the action of cellular enzymes on the hydroxylamine products initially generated by NfsA or NfsB. Relatively little of the 4-NHOH and derived 4-NH2 species were detectable with SKOV3-NfsA cells; the 2-NO2 reduction products were in 25- to 40-fold excess over the products of 4-NO2 reduction (at 30 min to 2 h). In contrast, with cells expressing NfsB, the levels of 2-NO2 and 4-NO2 reduction products were more similar (only ∼1.7- to 1.8-fold excess of 2-NO2 metabolites, at 1–2 h) (Figures 4E, F). The departure from a 1 : 1 ratio of 2-NO2 and 4-NO2 reduction products released to the medium by SKOV3-NfsB cells may be attributable to greater reactivity of the 4-NHOH species inside the cell. Metabolite ratios were similar in a separate experiment using a more pharmacologically relevant CB1954 concentration (10 μM), which confirmed the preference of NfsA for the reduction of the 2-NO2 group (data not shown). Thus, in cells and with the purified enzyme, NfsA shows a strong bias towards reduction of the 2-NO2 group of CB1954.
Figure 4.
HPLC analysis of time course and regiospecificity of CB1954 reduction by SKOV3 cells expressing NfsA or NfsB. (A, C, E) Time courses of CB1954 reduction and accumulation of CB1954 metabolites in extracellular medium (mean and s.e.m of three experiments; key in panel A also applies for panels C and E); no metabolites of CB1954 were detected using SKOV3-GFP cells (panel A). (B, D, F) HPLC traces of medium sampled after 2 h incubation. Peaks marked C and P correspond to CB1954 and phenol red; peaks marked 2 and 4 correspond to the 2-NHOH and 4-NHOH products and those marked 2′ and 4′ indicate the corresponding -NH2 metabolites. Panels A and B represent SKOV3-GFP cells (control); panels C and D represent SKOV3-NfsA cells; and panels E and F represent SKOV3-NfsB cells.
Bystander effect of NfsA nitroreductase
Helsby et al (2004) suggested that the bystander effect from the activation of CB1954 is largely caused by the 2-NHOH and derived 2-NH2 metabolites, rather than the 4-NHOH product. Because NfsA nitroreductase preferentially reduces the 2-NO2 group of CB1954 as shown above, we anticipated that it might produce a greater bystander effect than NfsB nitroreductase. The stably transduced cell lines, SKOV3-NfsA and SKOV3-NfsB, are closely matched for sensitivity to CB1954 (as shown in Figure 5A and Table 2 ), facilitating comparison of the bystander effect with the two enzymes. Untransduced ‘target’ SKOV3 cells and transduced ‘activator’ (SKOV3-NfsA or SKOV3-NfsB) cells were mixed in various proportions before testing the susceptibility of the mixed cultures to CB1954. Figure 5A shows a selection of the dose-response curves for cell survival in the mixed cultures. The IC50 (CB1954 concentration giving 50% reduction in viable cells) for SKOV3 cells alone was 400 μM CB1954, whereas IC50s for cultures of 100% SKOV3-NfsA or SKOV3-NfsB were ∼0.6 μM CB1954. The mixed cell populations showed CB1954 dose-response curves and IC50s intermediate between those of pure activator and pure target cells, depending on the proportion of activator cells and the enzyme (NfsA or NfsB). The IC50s for all cell mixtures, and the unmixed cells, are plotted against percentage activator cells in Figure 5B. The IC50s for mixtures with SKOV3-NfsA cells are lower than for mixtures of SKOV3-NfsB cells over the entire range from 1.56 to 50% activator cells. For mixtures containing between 3.12 and 25% activator cells, this advantage equates to a three- to five-fold lower IC50 when using NfsA. Conversely, if we consider the proportion of activator cells required to achieve an IC50 of 15 μM CB1954 (midway on a log scale between the IC50 of SKOV3 cells and either activator cell type), this would require 13.7% of SKOV3-NfsB cells, but only 5.7% of SKOV3-NfsA cells (P<0.01).
Figure 5.
Bystander effect with NfsA vs NfsB. (A) ‘Activator cells’, that is, SKOV3 cells stably expressing either NfsA or NfsB, parental SKOV3 cells and cell mixtures containing 50, 25, 12.5, 6.25, 3.12 and 1.56% of the activator cells mixed with SKOV3 target cells, were exposed to a range of CB1954 concentrations, and their subsequent survival determined using the MTT assay; symbols show the mean and range of duplicate wells. For clarity, only dose-response curves for 25 and 6.25% activator cells are shown. (B) The IC50s (±95% CI), derived from the MTT assay cell survival curves in panel A are plotted against percentage (%) activator cells in the mixture. The horizontal lines indicate the IC50s of parental SKOV3 cells (upper line), pure cultures of SKOV3-NfsA and SKOV3-NfsB (lower line), and the prodrug concentration midway between these on a log scale.
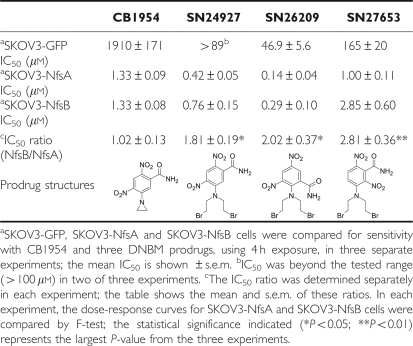
Table 2. Improved cell sensitization to DNBM prodrugs using NfsA vs NfsB.
Comparison of NfsA and NfsB nitroreductase for activation of alternative prodrugs
The nfsA and nfsB genes were originally identified through their role in sensitising bacteria to the nitrofuran antibiotics, nitrofurazone and nitrofurantoin. NfsB has been shown to sensitise human cells to nitrofurazone (Bailey et al, 1996). We compared the two enzymes using SKOV3 cells infected with 50 p.f.u. per cell of CTL102 or AdSV042. Mock-infected cells had IC50s of 410 and 520 μM nitrofurazone and nitrofurantoin, respectively. The expression of NfsB by CTL102 resulted in 23- and 17-fold sensitisation, respectively, whereas that of NfsA by AdSV042 caused 71- and 34-fold sensitisation, indicating that NfsA is approximately three- and two-fold better than NfsB for cell sensitisation to nitrofurazone and nitrofurantoin, respectively. However, in cell mixing experiments, neither of these prodrugs gave detectable bystander cell killing, using either NfsA or NfsB (data not shown).
The DNBMs are another promising class of prodrug activated by NfsB (Anlezark et al, 1995; Wilson et al, 2002). We evaluated the ability of NfsA to activate three representative DNBM prodrugs using the SKOV3-NfsA and SKOV3-NfsB cell lines. As shown in Table 2, although these cell lines were matched for equal sensitivity to CB1954, the IC50s for all three DNBM prodrugs were ∼1.8- to 2.8-fold lower for SKOV3-NfsA than for SKOV3-NfsB cells, indicating that NfsA also activates this family of prodrugs more efficiently than NfsB.
Discussion
We have shown that NfsA nitroreductase can activate CB1954 significantly faster than NfsB at low substrate concentrations when using the NADPH cofactor, and that it selectively reduces the 2-NO2 rather than the 4-NO2 group (whereas NfsB shows no preference). The improved kinetics may result from the more open active site of NfsA (Kobori et al, 2001; Lovering et al, 2001), in particular, the absence of any amino acids in NfsA at a position corresponding to the residue Phe124 of NfsB, mutation of which to a wide variety of alternative residues improved the rate of CB1954 activation (Grove et al, 2003). However, a more detailed explanation of the differences in substrate binding, in particular to explain the regioselectivity of CB1954 activation by NfsA, will require further studies. We have identified two explanations regarding why the initial purification and cloning of an enzyme capable of CB1954 activation from E. coli isolated NfsB rather than NfsA (Anlezark et al, 1992). First, the assay used to monitor purification used the cofactor NADH, whereas NfsA has a strong preference for NADPH (see Figure 2) (Bryant et al, 1981; Zenno et al, 1996); furthermore, this assay used menadione as a substrate, which is reduced about twice as efficiently by NfsB as by NfsA (Zenno et al, 1996). Although the initial demonstration that NfsB could activate CB1954 (and does so ∼90-fold faster than rat DT-diaphorase (Anlezark et al, 1992)) led to the adoption of NfsB for prodrug activation gene therapy with CB1954, this paper presents evidence based on cell cultures that it may be advantageous to use NfsA in preference.
For an enzyme reaction at substrate concentrations far below the Km will be the case with either NfsA or NfsB at the concentrations of CB1954 achievable in patients (<10 μM (Chung-Faye et al, 2001)), the rate of reaction is proportional to kcat/Km. With purified enzymes in vitro, this ratio was 25-fold higher for NfsA than for NfsB; taking into account a modest differential inhibition of the two enzymes by DMSO in the reaction buffer, the data indicated that NfsA is ∼18-fold more efficient at reducing CB1954. In agreement with this, when using the NADPH cofactor, purified NfsA was ∼20-fold more potent than NfsB (with either cofactor) for sensitising SKOV3 ovarian carcinoma cells to CB1954. The relative rates of CB1954 activation by NfsA and NfsB expressed within cells will depend on the intracellular concentrations of NADH and NADPH. The maximum benefit of NfsA would occur if the intracellular NADPH concentration greatly exceeded that of NADH; conversely as indicated by Figure 3A, a preponderance of NADH would favour NfsB. We could find very few reports of cofactor concentrations in cells or tissues; one study measured approximately 15–44 μM NADPH in three B-cell lines (Hancock et al, 1989), and HPLC analysis of HEK293 cells (adenovirus-transformed human embryonic kidney cells) showed NADH levels that appeared approximately 1–2 times greater than NADPH (Pollak et al, 2007). To compare cancer cell sensitisation with CB1954 by the enzymes after gene transfer, we used replication-defective adenoviruses AdSV042 (expressing NfsA) compared with the similar adenovirus, CTL102 (expressing NfsB). In three experiments, the adenovirus expressing NfsA achieved ∼3.5-, 6.3- and 8.1-fold lower IC50 than that expressing NfsB (comparisons at 300 p.f.u. per cell; P<0.001 for all differences). In the absence of available antibodies capable of detecting NfsA, we cannot independently confirm that NfsA and NfsB are expressed at equal levels in the target cells; however, as they were delivered and expressed by the otherwise identical adenovirus vectors, these experiments provide a fair indication of their relative potential benefit for prodrug activation gene therapy. The reduced benefit observed with intracellularly expressed NfsA, compared with extracellular enzymes with added NADPH, might be explained by the available concentrations of NADH and NADPH within the cells.
We observed that NfsA preferentially reduces the 2-NO2 rather than the 4-NO2 group of CB1954 to the hydroxylamine (−NHOH), whereas NfsB generates equal amounts of the 2-NHOH and 4-NHOH products. Studies of the diffusion properties of CB1954 metabolites through multicellular layers (Helsby et al, 2004) suggested that the 2-NO2 reduction products should be more effective at bystander cell killing. In cell mixing experiments with various proportions of ‘activator’ cells expressing NfsA or NfsB, we confirmed that the expression of NfsA conferred a greater bystander effect with CB1954 than was obtained with NfsB.
We have also shown that expressing NfsA rather than NfsB from an adenovirus vector is more efficient for sensitising human cells to nitrofurazone and nitrofurantoin. Although the therapeutic index of these nitrofuran antibiotics is lower than for CB1954, they have the advantage of established high clinical tolerability. The lack of significant bystander cell killing with these prodrugs would limit their utility for GDEPT. Nevertheless, NfsA may be useful for cell sensitisation to nitrofurazone or nitrofurantoin, for applications in which bystander cell killing may be undesirable—for example, ablation of adoptively transferred cells or selective cell or tissue ablation in transgenic animals (Isles et al, 2001).
Another promising class of alternative prodrugs activated by NfsB are the DNBMs (Anlezark et al, 1995; Atwell et al, 2007). SN24927 is the bromo-analogue of SN23862, the most studied alternative prodrug to CB1954 for use in combination with NfsB nitroreductase (Helsby et al, 2004). Earlier studies using NfsB expression to activate the 2,4-dinitrobenzamide-5-mustard, SN24927, showed a greater bystander effect than with CB1954, and superior anti-tumour activity in mice (Wilson et al, 2002; Atwell et al, 2007). A 3,5-dinitrobenzamide-2-mustard closely related to SN26209 (Table 2) has been shown recently to be a potent NfsB prodrug (Singleton et al, 2007). All DNBM prodrugs require reduction of the nitro group para to the mustard for activation; this is equivalent to the 2-NO2 group of CB1954, which we have shown to be preferentially reduced by NfsA. Using SKOV3 cells expressing NfsA or NfsB and matched for sensitivity to CB1954, we found that the SKOV3-NfsA cells were 1.8- to 2.8-fold more sensitive to the DNBM prodrugs than SKOV3-NfsB, indicating that the benefit of using NfsA rather than NfsB to activate these prodrugs is greater than with CB1954.
An earlier screen of different bacteria for nitroreductases suitable for prodrug activation therapies identified YwrO from Bacillus amyloliquefaciens, which selectively reduces the 4-NO2 group of CB1954 and was reported to have a modest kinetic advantage relative to E. coli NfsB (Anlezark et al, 2002). However, YwrO seemed markedly less effective than NfsB at sensitising V79 hamster cells to CB1954, and was unable to activate the DNBM prodrug SN23862. Another enzyme from E. coli, YieF, was shown to be comparable with NfsA for sensitising human cells to CB1954, although neither was compared with NfsB (Barak et al, 2006). As the cytotoxicity of at least the 4-hydroxylamino derivative of CB1954 was proposed to require acetylation (Knox et al, 1991), the possible benefit of co-expressing acetyl transferases with NfsB has been tested. Co-expression of human N-acetyl transferase-2 was shown to significantly enhance the sensitisation of NfsB-expressing cells to CB1954 (Mitchell and Minchin, 2008); however, the bystander effect was concomitantly reduced.
The 3.5- to 8.1-fold improvement in the sensitisation of human cells to CB1954 by adenovirus vectors expressing NfsA relative to NfsB is similar to the level of improvement that has been achieved by the best single amino acid substitutions within the active site of NfsB (Grove et al, 2003). Further multiple-substitution mutants of NfsB further optimised for CB1954 activation have been selected (Guise et al, 2007), and mutants of YieF showing improved sensitisation to CB1954 relative to NfsA were reported (Barak et al, 2006). Nevertheless, the observed improvement in bystander cell killing attributable to preferential reduction of the 2-NO2 group of CB1954 by NfsA is a desirable feature, suggesting that NfsA could itself be a good starting enzyme for further mutagenic enhancement of its catalytic rate. This regioselectivity of nitroreduction by NfsA is also reflected in its improved cell sensitisation to DNBM prodrugs relative to NfsB. We conclude that NfsA should be considered as an alternative prodrug-activating enzyme to NfsB for use in prodrug activation gene therapy or cell/tissue ablation studies using CB1954, nitrofuran antibiotics or DNBM prodrugs.
Acknowledgments
We thank Dianne Ferry and Graham Burns for their assistance with HPLC analysis of cellular or in vitro metabolites of CB1954, respectively. We also thank Graham Atwell and Professor WA Denny for supply of the DNBM compounds. SOV was in receipt of a PhD studentship from the MRC and a Universitas 21 PhD Travel Scholarship, and also received support from Cancer Research UK (Grant No. C1007/A6688). DJ was funded by a BBSRC CASE studentship, partnered with ML Laboratories plc.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Anlezark GM, Melton RG, Sherwood RF, Coles B, Friedlos F, Knox RJ (1992) The bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954)–I Purification and properties of a nitroreductase enzyme from Escherichia coli–a potential enzyme for antibody-directed enzyme prodrug therapy (ADEPT). Biochem Pharmacol 44: 2289–2295 [DOI] [PubMed] [Google Scholar]
- Anlezark GM, Melton RG, Sherwood RF, Wilson WR, Denny WA, Palmer BD, Knox RJ, Friedlos F, Williams A (1995) Bioactivation of dinitrobenzamide mustards by an E coli B nitroreductase. Biochem Pharmacol 50: 609–618 [DOI] [PubMed] [Google Scholar]
- Anlezark GM, Vaughan T, Fashola-Stone E, Paul Michael N, Murdoch H, Sims MA, Stubbs S, Wigley S, Minton NP (2002) Bacillus amyloliquefaciens orthologue of Bacillus subtilis ywrO encodes a nitroreductase enzyme which activates the prodrug CB 1954. Microbiology 148: 297–306 [DOI] [PubMed] [Google Scholar]
- Atwell GJ, Yang S, Pruijn FB, Pullen SM, Hogg A, Patterson AV, Wilson WR, Denny WA (2007) Synthesis and structure-activity relationships for 2,4-Dinitrobenzamide-5-mustards as prodrugs for the Escherichia coli nfsB nitroreductase in gene therapy. J Med Chem 50(6): 1197–212 [DOI] [PubMed] [Google Scholar]
- Bailey SM, Knox RJ, Hobbs SM, Jenkins TC, Mauger AB, Melton RG, Burke PJ, Connors TA, Hart IR (1996) Investigation of alternative prodrugs for use with E coli nitroreductase in ‘suicide gene’ approaches to cancer therapy. Gene Ther 3: 1143–1150 [PubMed] [Google Scholar]
- Barak Y, Thorne SH, Ackerley DF, Lynch SV, Contag CH, Matin A (2006) New enzyme for reductive cancer chemotherapy, YieF, and its improvement by directed evolution. Mol Cancer Ther 5: 97–103 [DOI] [PubMed] [Google Scholar]
- Bridgewater JA, Springer CJ, Knox RJ, Minton NP, Michael NP, Collins MK (1995) Expression of the bacterial nitroreductase enzyme in mammalian cells renders them selectively sensitive to killing by the prodrug CB1954. Eur J Cancer 31A: 2362–2370 [DOI] [PubMed] [Google Scholar]
- Bryant DW, McCalla DR, Leeksma M, Laneuville P (1981) Type I nitroreductases of Escherichia coli. Can J Microbiol 27: 81–86 [DOI] [PubMed] [Google Scholar]
- Chung-Faye G, Palmer D, Anderson D, Clark J, Downes M, Baddeley J, Hussain S, Murray PI, Searle P, Seymour L, Harris PA, Ferry D, Kerr DJ (2001) Virus-directed, enzyme prodrug therapy with nitroimidazole reductase: a phase I and pharmacokinetic study of its prodrug, CB1954. Clin Cancer Res 7: 2662–2668 [PubMed] [Google Scholar]
- Cosset FL, Takeuchi Y, Battini JL, Weiss RA, Collins MK. (1995) High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol 69: 7430–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djeha AH, Hulme A, Dexter MT, Mountain A, Young LS, Searle PF, Kerr DJ, Wrighton CJ (2000) Expression of Escherichia coli B nitroreductase in established human tumor xenografts in mice results in potent antitumoral and bystander effects upon systemic administration of the prodrug CB1954. Cancer Gene Ther 7: 721–731 [DOI] [PubMed] [Google Scholar]
- Djeha AH, Thomson TA, Leung H, Searle PF, Young LS, Kerr DJ, Harris PA, Mountain A, Wrighton CJ (2001) Combined adenovirus-mediated nitroreductase gene delivery and CB1954 treatment: a well-tolerated therapy for established solid tumors. Mol Ther 3: 233–240 [DOI] [PubMed] [Google Scholar]
- Fallaux FJ, Kranenburg O, Cramer SJ, Houweling A, van Ormondt H, Hoeben RC, van der Eb AJ (1996) Characterization of 911: a new helper cell line for the titration and propagation of early-region-1-deleted adenoviral vectors. Hum Gene Ther 7: 215–222 [DOI] [PubMed] [Google Scholar]
- Freeman SM, Abboud CN, Whartenby KA, Packman CH, Koeplin DS, Moolten FL, Abraham GN (1993) The bystander effect: tumor regression when a fraction of the tumor mass is genetically modified. Cancer Res 53: 5274–5283 [PubMed] [Google Scholar]
- Friedlos F, Quinn J, Knox RJ, Roberts JJ (1992) The properties of total adducts and interstrand crosslinks in the DNA of cells treated with CB 1954. Biochem Pharmacol 43: 1249–1254 [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ (1973) Transformation of rat cells by DNA of human adenovirus 5. Virology 54: 536–539 [DOI] [PubMed] [Google Scholar]
- Green NK, Youngs DJ, Neoptolemos JP, Friedlos F, Knox RJ, Springer CJ, Anlezark GM, Michael NP, Melton RG, Ford MJ, Young LS, Kerr DJ, Searle PF (1997) Sensitization of colorectal and pancreatic cancer cell lines to the prodrug 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954) by retroviral transduction and expression of the E coli nitroreductase gene. Cancer Gene Ther 4: 229–238 [PubMed] [Google Scholar]
- Grove JI, Lovering AL, Guise C, Race PR, Wrighton CJ, White SA, Hyde EI, Searle PF (2003) Generation of Escherichia coli nitroreductase mutants conferring improved cell sensitization to the prodrug CB1954. Cancer Res 63: 5532–5537 [PubMed] [Google Scholar]
- Grove JI, Searle PF, Weedon SJ, Green NK, McNeish IA, Kerr DJ (1999) Virus directed enzyme prodrug therapy using CB1954. Anticancer Drug Des 14: 461–472 [PubMed] [Google Scholar]
- Guise CP, Grove JI, Hyde EI, Searle PF (2007) Direct positive selection for improved nitroreductase variants using SOS triggering of bacteriophage lambda lytic cycle. Gene Ther 14: 690–698 [DOI] [PubMed] [Google Scholar]
- Hancock JT, Maly FE, Jones OT (1989) Properties of the superoxide-generating oxidase of B-lymphocyte cell lines. Determination of Michaelis parameters. Biochem J 262: 373–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsby NA, Ferry DM, Patterson AV, Pullen SM, Wilson WR (2004) 2-Amino metabolites are key mediators of CB 1954 and SN 23862 bystander effects in nitroreductase GDEPT. Br J Cancer 90: 1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsby NA, Wheeler SJ, Pruijn FB, Palmer BD, Yang S, Denny WA, Wilson WR (2003) Effect of nitroreduction on the alkylating reactivity and cytotoxicity of the 2,4-Dinitrobenzamide-5-aziridine CB 1954 and the corresponding nitrogen mustard SN 23862: distinct mechanisms of bioreductive activation. Chem Res Toxicol 16: 469–478 [DOI] [PubMed] [Google Scholar]
- Isles AR, Ma D, Milsom C, Skynner MJ, Cui W, Clark J, Keverne EB, Allen ND (2001) Conditional ablation of neurones in transgenic mice. J Neurobiol 47: 183–193 [DOI] [PubMed] [Google Scholar]
- Knox RJ, Friedlos F, Jarman M, Roberts JJ (1988) A new cytotoxic, DNA interstrand crosslinking agent, 5-(aziridin-1-yl)-4-hydroxylamino-2-nitrobenzamide, is formed from 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) by a nitroreductase enzyme in Walker carcinoma cells. Biochem Pharmacol 37: 4661–4669 [DOI] [PubMed] [Google Scholar]
- Knox RJ, Friedlos F, Marchbank T, Roberts JJ (1991) Bioactivation of CB 1954: reaction of the active 4-hydroxylamino derivative with thioesters to form the ultimate DNA-DNA interstrand crosslinking species. Biochem Pharmacol 42: 1691–1697 [DOI] [PubMed] [Google Scholar]
- Knox RJ, Friedlos F, Sherwood RF, Melton RG, Anlezark GM (1992) The bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954)—II Journal title: a comparison of an Escherichia coli nitroreductase and Walker DT diaphorase. Biochem Pharmacol 44: 2297–2301 [DOI] [PubMed] [Google Scholar]
- Kobori T, Sasaki H, Lee WC, Zenno S, Saigo K, Murphy ME, Tanokura M (2001) Structure and site-directed mutagenesis of a flavoprotein from Escherichia coli that reduces nitrocompounds: alteration of pyridine nucleotide binding by a single amino acid substitution. J Biol Chem 276: 2816–2823 [DOI] [PubMed] [Google Scholar]
- Lovering AL, Hyde EI, Searle PF, White SA (2001) The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7Å, 1.8Å and 2.4Å resolution. J Mol Biol 309: 203–213 [DOI] [PubMed] [Google Scholar]
- McNeish IA, Green NK, Gilligan MG, Ford MJ, Mautner V, Young LS, Kerr DJ, Searle PF (1998) Virus directed enzyme prodrug therapy for ovarian and pancreatic cancer using retrovirally delivered E coli nitroreductase and CB 1954. Gene Ther 5: 1061–1069 [DOI] [PubMed] [Google Scholar]
- McNeish IA, Searle PF, Young LS, Kerr DJ (1997) Gene directed enzyme prodrug therapy for cancer. Adv Drug Deliv Rev 26: 173–184 [DOI] [PubMed] [Google Scholar]
- Mitchell DJ, Minchin RF (2008) E coli nitroreductase/CB 1954 gene directed enzyme prodrug therapy: role of arylamine N-acetlytransferase 2. Cancer Gene Ther 15: 758–764 [DOI] [PubMed] [Google Scholar]
- Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65: 55–63 [DOI] [PubMed] [Google Scholar]
- Niculescu-Duvaz I, Springer CJ (2005) Introduction to the background, principles, and state of the art in suicide gene therapy. Mol Biotechnol 30: 71–88 [DOI] [PubMed] [Google Scholar]
- Palmer DH, Mautner V, Mirza D, Oliff S, Gerritsen W, Van Der Sijp JR, Hubscher S, Reynolds G, Bonney S, Rajaratnam R, Hull D, Horne M, Ellis J, Mountain A, Hill S, Harris PA, Searle PF, Young LS, James ND, Kerr DJ (2004) Virus-directed enzyme prodrug therapy: intratumoral administration of a replication-deficient adenovirus encoding nitroreductase to patients with resectable liver cancer. J Clin Oncol 22: 1546–1552 [DOI] [PubMed] [Google Scholar]
- Patel P, Young JG, Mautner V, Ashdown D, Bonney S, Pineda RG, Collins SI, Searle PF, Hull D, Peers E, Chester J, Wallace DM, Doherty A, Leung H, Young LS, James ND (2009) A Phase I/II Clinical Trial in Localized Prostate Cancer of an Adenovirus Expressing Nitroreductase with CB1954. Mol Ther, doi:10.1038/mt.2009.80 [DOI] [PMC free article] [PubMed]
- Pollak N, Niere M, Ziegler M (2007) NAD kinase levels control the NADPH concentration in human cells. J Biol Chem 282: 33562–33571 [DOI] [PubMed] [Google Scholar]
- Race PR, Lovering AL, Green RM, Ossor A, White SA, Searle PF, Wrighton CJ, Hyde EI (2005) Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone: reversed binding orientations in different redox states of the enzyme. J Biol Chem 280: 13256–13264 [DOI] [PubMed] [Google Scholar]
- Race PR, Lovering AL, White SA, Grove JI, Searle PF, Wrighton CW, Hyde E (2007) Kinetic and structural characterisation of Escherichia coli nitroreductase mutants showing improved efficacy for the prodrug substrate CB1954. J Mol Biol 368: 481–492 [DOI] [PubMed] [Google Scholar]
- Russell PJ, Khatri A (2006) Novel gene-directed enzyme prodrug therapies against prostate cancer. Expert Opin Investig Drugs 15: 947–961 [DOI] [PubMed] [Google Scholar]
- Sastry SS, Jayaraman R (1984) Nitrofurantoin-resistant mutants of Escherichia coli: isolation and mapping. Mol Gen Genet 196: 379–380 [DOI] [PubMed] [Google Scholar]
- Searle PF, Chen MJ, Hu L, Race PR, Lovering AL, Grove JI, Guise C, Jaberipour M, James ND, Mautner V, Young LS, Kerr DJ, Mountain A, White SA, Hyde EI (2004) Nitroreductase: a prodrug-activating enzyme for cancer gene therapy. Clin Exp Pharmacol Physiol 31: 811–816 [DOI] [PubMed] [Google Scholar]
- Singleton DC, Li D, Bai SY, Syddall SP, Smaill JB, Shen Y, Denny WA, Wilson WR, Patterson AV (2007) The nitroreductase prodrug SN 28343 enhances the potency of systemically administered armed oncolytic adenovirus ONYX-411(NTR). Cancer Gene Ther 14: 953–967 [DOI] [PubMed] [Google Scholar]
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR (1990) New colorimetric cytotoxic assay for anticancer-drug screening. J Natl Cancer Inst 82: 1107–1112 [DOI] [PubMed] [Google Scholar]
- Weedon SJ, Green NK, McNeish IA, Gilligan MG, Mautner V, Wrighton CJ, Mountain A, Young LS, Kerr DJ, Searle PF (2000) Sensitisation of human carcinoma cells to the prodrug CB1954 by adenovirus vector-mediated expression of E coli nitroreductase. Int J Cancer 86: 848–854 [DOI] [PubMed] [Google Scholar]
- Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert IB (1998) Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol 180: 5529–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WR, Pullen SM, Hogg A, Helsby NA, Hicks KO, Denny WA (2002) Quantitation of bystander effects in nitroreductase suicide gene therapy using three-dimensional cell cultures. Cancer Res 62: 1425–1432 [PubMed] [Google Scholar]
- Zenno S, Koike H, Kumar AN, Jayaraman R, Tanokura M, Saigo K (1996) Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol 178: 4508–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.