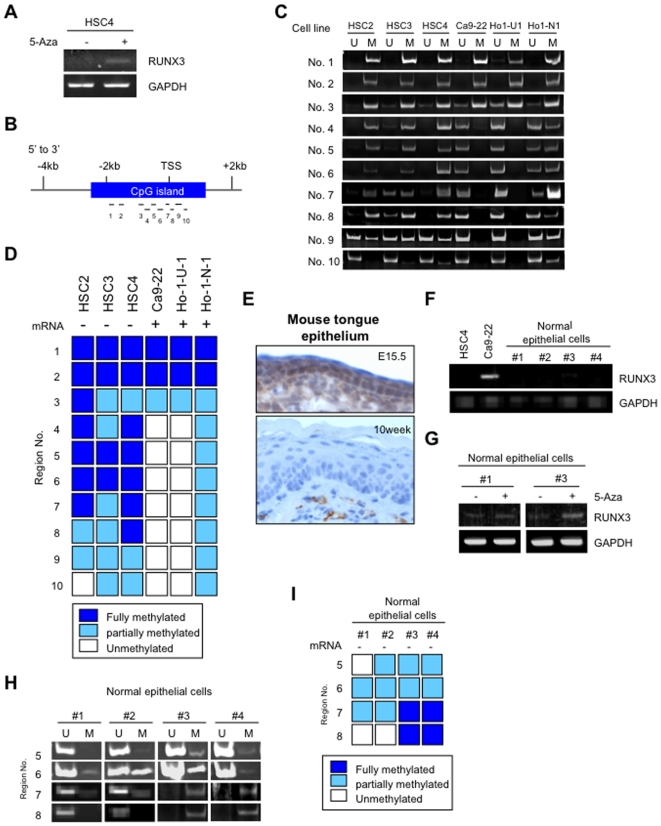
Figure 3. Methylation status in normal oral epithelial cells and HNSCC.
A: RUNX3 expression was examined after 5-aza-2′-deoxycytidine (5-Aza) treatment. HSC4 cells were treated with medium containing 300 nM 5-aza-2′-deoxycytidine for 72 h. After treatment, cells were collected and examined the expression of RUNX3 mRNA by RT-PCR. B: RUNX3 CpG island and analyzed regions (No. 1–10) are shown as vertical bars. Transcriptional start site (TSS) is located within region No. 7. C: Methylation status of RUNX3 in HNSCC cell lines. Genomic DNA was extracted from HNSCC cell lines and was treated with bisulfite. Methylation status of promoter region (No. 1–10) was examined by methylation specific PCR method. D: The summary of methylation and expression status of RUNX3 in HNSCC cell lines. E: Expression of Runx3 in mouse tongue epithelium of embryo (upper panel) and adult (lower panel) mouse by immunohistochemistry. Tongue tissues of mouse embryos at embryonic day 15.5 and 10 weeks old BALB/c mice were used. F: RUNX3 mRNA expression was examined by RT-PCR in 4 primary cultured normal oral epithelial cells (#1–#4). HSC4 cell was used as a negative control and Ca9-22 cell was used as a positive control for RUNX3 expression. GAPDH was used as a control. G: RUNX3 expression was examined after 5-aza-2′-deoxycytidine (5-Aza) treatment. Primary cultured normal oral epithelial cells (#1 and #2) were treated with medium containing 300 nM 5-aza-2′-deoxycytidine for 72 h. After treatment, cells were collected and examined the expression of RUNX3 mRNA by RT-PCR. H: Genomic DNA was extracted from 4 primary cultured normal oral epithelial cells (#1–#4). Methylation status at region No. 5–8 was examined by methylation specific PCR method. I: The summary of methylation and expression status of RUNX3 in normal epithelial cells.