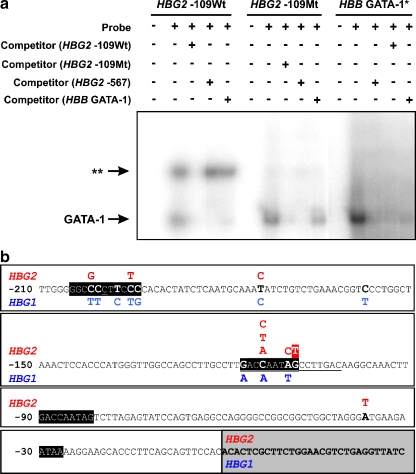
Fig. 3.
a Electrophoretic mobility shift assays of the oligonucleotides containing either the wild-type (Wt; HBG2:g-109 G) or mutant (Mt; HBG2:g-109 T) sequence using nuclear protein extracts prepared from dimethyl sulfoxide-induced MEL cells. The HBG2 −109Mt oligonucleotide most likely abolishes a NF-E3 binding site in vitro (depicted as two asterisks). Competition with nonlabeled HBG2 −109Wt oligonucleotide results in the disappearance of GATA-1 and NF-E3 band shifts, suggesting that both proteins bind to the HBG2 −109Wt oligonucleotide. Competition with nonlabeled HBG2 −567 or HBB GATA-1 oligonucleotides [15] results in the disappearance only of the GATA-1 (lower) band. Competition with nonlabeled HBG2 −567 or HBB GATA-1 oligonucleotides suggests that the affinity of GATA-1 is stronger to the former oligonucleotide. Use of nuclear protein extracts prepared from K562 cells yielded identical electrophoretic mobility shifts (not shown). The asterisk indicates the oligonucleotide sequence located in the HBB gene promoter region. b Schematic representation of the various nd-HPFH mutations reported to date for the HBG2 (in red) and HBG1 (in blue) globin genes (underlined sequences depict the two small deletions also leading to nd-HPFH). The novel nd-HPFH mutation reported in this study is highlighted in red. Gray box represents the sequences downstream to the transcription initiation site and sequences in solid black boxes represent phylogenetically conserved fetal globin genes cis-regulatory elements