Abstract
Wild birds of the orders Anseriformes and Charadriiformes are the natural reservoirs for avian influenza (AI) viruses. Traditionally, AI virus surveillance in wild birds has relied on virus identification strategies, including virus isolation and detection. To evaluate the accuracy of a commercial blocking enzyme-linked immunosorbent assay (bELISA) and the agar gel immunodiffusion (AGID) test for detection of antibodies in wild birds, which is indicative of AI virus infection, we tested 281 serum samples from various wild avian species that were experimentally infected with AI viruses. Included in these samples were 178 samples from birds with confirmed AI virus infections (122 infected with low-pathogenic AI [LPAI] viruses and 56 infected with highly pathogenic AI [HPAI] viruses) and 103 samples from birds that were uninfected, negative controls. The sensitivities of the bELISA and the AGID test were 0.820 (95% confidence interval [95% CI], 0.756 to 0.874) and 0.674 (95% CI, 0.600 to 0.742), respectively. Both tests had an estimated specificity of 1.00 (95% CI, 0.965 to 1.00). The bELISA was significantly more sensitive than the AGID test for both LPAI virus- and HPAI virus-infected birds. Both assays, however, had a higher sensitivity for birds infected with HPAI virus than for birds infected with LPAI virus. These results demonstrate the potential utility of the bELISA for detection of antibodies to both LPAI and HPAI viruses in multiple avian species, representing five avian orders and 17 genera. Additional studies are warranted to further evaluate the utility of the bELISA for use with naturally infected birds.
Avian influenza (AI) viruses have been reported from a wide diversity of free-living birds representing over 100 species in 12 taxonomic orders (11). All of the known hemagglutinin (H1 to H16) and neuraminidase (N1 to N9) subtypes of AI viruses have been isolated from wild birds (8, 9, 11), and currently, species in the orders Anseriformes (ducks, geese, and swans) and Charadriiformes (gulls, terns, and shorebirds) are believed to be the natural reservoirs. Surveillance for AI virus in wild-bird populations is predominately dependent on diagnostic assays that identify the virus, including reverse transcriptase PCR and virus isolation. Virus isolation from oropharyngeal or cloacal swabs of embryonating chicken eggs currently represents the preferred method of AI virus diagnosis and surveillance in wild-bird populations. However, agent-specific identification assays, such as virus isolation and reverse transcriptase PCR, are expensive, labor-intensive, and dependent on the host actively excreting virus. Consequently, a limitation of the agent identification-based approach to AI virus surveillance in wild birds relates to the relatively short duration of viral shedding and the high degree of spatial and temporal variations in viral prevalence within different wild avian populations. These limitations and uncertainties often necessitate large sample sizes to identify positives and repeat sampling at different times and locations. Additionally, the variability creates difficulty when interpreting negative test results, i.e., in determining whether a negative result is indicative of inappropriate sampling (wrong location or time) or a species that is resistant to or rarely infected with AI virus.
Serologic assays are commonly utilized for surveillance and diagnostics with domestic poultry to detect whether a population of birds has previously been exposed to an AI virus. Serologic tests utilized for AI virus antibody detection in domestic poultry include the agar gel immunodiffusion (AGID) test, the enzyme-linked immunosorbent assay (ELISA), the hemagglutination inhibition test, and the neuraminidase inhibition test (17). The AGID test and the ELISA detect antibodies against all type A influenza viruses and consequently are the preferred assays for use as a screening tool. The hemagglutination inhibition and the neuraminidase inhibition tests are hemagglutinin and neuraminidase specific, respectively, and typically are performed to identify antibodies to specific subtypes or to confirm AGID test- or ELISA-positive samples when information on the subtype is available.
The AGID test is the most commonly utilized serologic assay for AI virus surveillance in domestic poultry and detects antibodies directed against the AI virus internal proteins nucleoprotein (NP) and matrix 1 (M1) protein (20). While the AGID test is inexpensive and simple to perform, the primary disadvantage is that it is only moderately sensitive for gallinaceous poultry (17). The sensitivities of the AGID test for nongalliform birds vary between different species (17), but the test reportedly lacks sensitivity for waterfowl (5, 21), even under experimental conditions (15). Presumably, the low sensitivity of the AGID test for waterfowl is due to a combination of reduced antibody responsiveness and deficiencies in measurable antibody functions dependent on bi- or multivalency, such as precipitation and hemagglutination (7).
The indirect ELISA (iELISA) provides several advantages over the AGID test, including more-rapid and -objective test results, as well as the ability to screen large numbers of samples in a semiautomated manner. The iELISA is generally more sensitive than the AGID test for chickens (16), but most commercially available iELISAs were developed specifically for use with poultry species. These assays utilize anti-chicken/turkey immunoglobulin G secondary antibodies for type A influenza virus antibody detection and are not designed to detect antibodies in other avian hosts. Recently, however, several commercially available blocking ELISAs (bELISAs) that utilize a mouse monoclonal antibody to compete with the host antibodies in the test serum for binding to the test plate antigen have been developed. Consequently, these assays should perform accurately with any avian species, but to date there are little data on the accuracy of these tests with nongalliform birds.
Over the last 5 to 10 years, an enormous amount of research has been dedicated to understanding AI virus transmission and maintenance in wild-bird populations. A sensitive and specific serologic assay to detect AI virus antibodies in wild birds would complement existing agent identification wild-bird surveillance strategies and potentially provide a cost-efficient, safe, and less time-sensitive approach to aid in the identification of wild-bird species that may be involved in the epidemiology of AI viruses. Such an assay would enable AI virus surveillance programs to refocus limited surveillance and research funds for virus detection and isolation to avian species with evidence of AI virus infection. The objective of this study was to evaluate the accuracy of a commercially available bELISA for detection of AI virus antibodies in multiple avian species experimentally infected with these viruses.
MATERIALS AND METHODS
Serum samples.
Avian serum samples utilized in this study (Table 1) were obtained from previous AI virus experimental-infection trials conducted at the Southeast Poultry Research Laboratory, United States Department of Agriculture (USDA), Agricultural Research Service (ARS), Athens, GA, or at the University of Georgia, Athens, GA (1, 2, 3, 4, 12, 13, 14, 23; D. Swayne, unpublished data; D. E. Stallknecht, unpublished data). A total of 281 serum samples were included in this study: 178 samples from birds with a confirmed AI virus infection, and 103 samples from uninfected, sham-inoculated birds that served as negative controls. Birds inoculated with an AI virus were considered to have a confirmed infection if AI virus was isolated from an oropharyngeal swab on or after 2 days postinoculation (dpi) or if virus was isolated from a cloacal swab on any day postinoculation. Birds excreting virus via the oropharynx on 1 dpi only were not considered infected, as this result could be due to residual inoculum; therefore, serum samples from such birds were not included in this analysis. Of the 178 samples from birds with a confirmed infection, 122 serum samples were from birds that were inoculated with a low-pathogenic AI (LPAI) virus, and 56 were from birds inoculated with a highly pathogenic AI (HPAI) virus. All of the sham-inoculated birds had negative virus isolation results and negative pre- and post-AGID test results. The experimental designs varied slightly between trials, but all serum samples were collected at 10 to 21 days after intranasal inoculation with either a sham inoculum or an AI virus. The majority of virus-exposed birds in each species listed in Table 1 were inoculated with high viral concentrations (≥105 median embryo infectious doses [EID50]); however, some individuals were infected with lower viral concentrations as part of dose-response trials, as follows: two house sparrows (Passer domesticus) were inoculated with 103.1 EID50 of A/whooper swan/Mongolia/244/05 (H5N1), one Japanese quail (Coturnix japonica) was inoculated with 103 EID50 of A/chicken/Alabama/75 (H4N8), one Pekin duck (Anas platyrhynchos) was inoculated with 103 EID50 of A/chicken/Alabama/75 (H4N8), and five Pekin ducks were inoculated with 103 EID50 of A/chicken/Alabama/75 (H4N8). After collection, all serum samples were stored at −20°C until the AGID tests were performed. The samples were returned to −20°C and thawed one additional time prior to testing with the bELISA.
TABLE 1.
Characteristics of avian serum samples used in this evaluation, including species of origin, viral inoculation treatment group, confirmed-infection status, and serologic assay results
| Order and vernacular name, species | Virus treatment (isolate[s])a | Sample sizeb | No. (%) of ELISA-positive samplesc/total no. of samples | Mean S/N ratio (range) | No. (%) of AGID test-positive samples/total no. of samples |
|---|---|---|---|---|---|
| Anseriformes | |||||
| Mallard, Anas platyrhynchos | HPAI (A, B, C) | 7 | 5/7 (71) | 0.33 (0.13-0.67) | 3/7 (43) |
| LPAI (L, M, N) | 52 | 48/52 (92) | 0.26 (0.08-0.81) | 47/52 (90) | |
| Sham | 29 | 0/29 (0) | 0.84 (0.58-1.10) | 0/29 (0) | |
| Pekin duck, Anas platyrhynchos | HPAI (G, H) | 11 | 11/11 (100) | 0.19 (0.13-0.26) | 11/11 (100) |
| LPAI (I, J) | 22 | 16/22 (73) | 0.41 (0.19-0.89) | 0/22 (0) | |
| Sham | 8 | 0/8 (0) | 0.91 (0.80-1.00) | 0/8 (0) | |
| Blue-winged teal, Anas discors | HPAI (A) | 2 | 2/2 (100) | 0.24 (0.08-0.40) | 1/2 (50) |
| Sham | 1 | 0/1 (0) | 0.61 | 0/1 (0) | |
| Northern pintail, Anas acuta | HPAI (A) | 1 | 1/1 (100) | 0.29 | 0/1 (0) |
| Sham | 1 | 0/1 (0) | 0.76 | 0/1 (0) | |
| Chiloe wigeon, Anas sibilatrix | Sham | 1 | 0/1 (0) | 0.58 | 0/1 (0) |
| Cinnamon teal, Anas cyanoptera | HPAI (D) | 3 | 3/3 (100) | 0.24 (0.14-0.41) | 2/3 (67) |
| LPAI (E) | 3 | 3/3 (100) | 0.31 (0.19-0.44) | 3/3 (100) | |
| Sham | 2 | 0/2 (0) | 0.92 (0.87-0.97) | 0/2 (0) | |
| Redhead, Aythya americana | HPAI (A, B) | 4 | 4/4 (100) | 0.12 (0.06-0.20) | 3/4 (75) |
| LPAI (L, M, N) | 12 | 7/12 (58) | 0.47 (0.17-1.05) | 7/12 (58) | |
| Sham | 5 | 0/5 (0) | 0.97 (0.82-1.24) | 0/5 (0) | |
| Wood duck, Aix sponsa | HPAI (A) | 5 | 5/5 (100) | 0.12 (0.05-0.21) | 5/5 (100) |
| LPAI (L, M, N) | 9 | 7/9 (78) | 0.40 (0.10-0.70) | 7/9 (78) | |
| Sham | 13 | 0/13 (0) | 1.05 (0.81-1.17) | 0/13 (0) | |
| Mandarin duck, Aix galericulata | HPAI (C) | 2 | 2/2 (100) | 0.19 (0.11-0.27) | 2/2 (100) |
| Sham | 2 | 0/2 (0) | 0.95 (0.95-0.96) | 0/2 (0) | |
| Ruddy shelduck, Tadorna ferruginea | Sham | 2 | 0/2 (0) | 0.98 (0.93-1.03) | 0/2 (0) |
| Cackling goose, Branta hutchinsii | HPAI (A) | 1 | 1/1 (100) | 0.17 | 1/1 (100) |
| Sham | 2 | 0/2 (0) | 0.95 (0.94-0.97) | 0/2 (0) | |
| Bar-headed goose, Anser indicus | HPAI (A) | 3 | 3/3 (100) | 0.20 (0.17-0.24) | 3/3 (100) |
| Sham | 2 | 0/2 (0) | 1.41 (0.95-1.88) | 0/2 (0) | |
| Greylag goose, Anser anser | HPAI (C) | 3 | 3/3 (100) | 0.15 (0.13-0.17) | 3/3 (100) |
| Sham | 1 | 0/1 (0) | 0.92 | 0/1 (0) | |
| Chinese goose, Anser cygnoides | LPAI (I, J, K) | 10 | 3/10 (30) | 0.62 (0.41-0.90) | 0/10 (0) |
| Whooper swan, Cygnus cygnus | Sham | 1 | 0/1 (0) | 1.14 | 0/1 (0) |
| Trumpeter swan, Cygnus buccinator | Sham | 1 | 0/1 (0) | 1.18 | 0/1 (0) |
| Mute swan, Cygnus olor | Sham | 3 | 0/3 (0) | 0.87 (0.86-0.88) | 0/3 (0) |
| Black swan, Cygnus atratus | Sham | 1 | 0/1 (0) | 1.10 | 0/1 (0) |
| Charadriiformes | |||||
| Herring gull, Larus argentatus | HPAI (A, B) | 4 | 4/4 (100) | 0.23 (0.20-0.30) | 4/4 (100) |
| Sham | 1 | 0/1 (0) | 1.08 | 0/1 (0) | |
| Laughing gull, Leucophaeus atricilla | HPAI (A) | 2 | 2/2 (100) | 0.23 (0.19-0.27) | 2/2 (100) |
| LPAI (L, M, N) | 10 | 7/10 (70) | 0.50 (0.25-1.17) | 9/10 (90) | |
| Sham | 9 | 0/9 (0) | 0.92 (0.80-1.17) | 0/9 (0) | |
| Galliformes | |||||
| Northern bobwhite, Colinus virginianus | Sham | 2 | 0/2 (0) | 1.00 (0.98-1.03) | 0/2 (0) |
| Chukar partridge, Alectoris chukar | HPAI (F) | 1 | 1/1 (100) | 0.13 | 1/1 (100) |
| Sham | 1 | 0/1 (0) | 0.90 | 0/1 (0) | |
| Japanese quail, Coturnix japonica | LPAI (I, K) | 4 | 1/4 (25) | 0.77 (0.48-0.95) | 1/4 (25) |
| Passeriformes | |||||
| Zebra finch, Taeniopygia guttata | Sham | 2 | 0/2 (0) | 1.36 (0.95-1.77) | 0/2 (0) |
| House finch, Carpodacus mexicanus | Sham | 1 | 0/1 (0) | 1.60 | 0/1 (0) |
| House sparrow, Passer domesticus | HPAI (A) | 2 | 2/2 (100) | 0.13 (0.11-0.15) | 2/2 (100) |
| Sham | 5 | 0/5 (0) | 1.20 (0.86-1.73) | 0/5 (0) | |
| European starling, Sturnus vulgaris | Sham | 2 | 0/2 (0) | 1.95 (1.70-2.21) | 0/2 (0) |
| Columbiformes | |||||
| Rock pigeon, Columba livia | HPAI (A) | 5 | 5/5 (100) | 0.29 (0.20-0.44) | 3/5 (60) |
| Sham | 5 | 0/5 (0) | 1.00 (0.86-1.09) | 0/5 (0) |
Virus isolates used were as follows: A, A/whooper swan/Mongolia/244/05 (H5N1); B, A/duck meat/Anyang/AVL-1/01 (H5N1); C, A/chicken/South Korea/IS/06 (H5N1); D, A/chicken/Chile/184240-1/02 (H7N3); E, A/chicken/Chile/176822/02 (H7N3); F, A/chicken/Hong Kong/220/1997 (H5N1); G, A/egret/Hong Kong/757.2/02 (H5N1); H, A/human/Prachinburi/6231/04 (H5N1); I, A/chicken/Alabama/75 (H4N8); J, A/mallard/Ohio/338/86 (H4N8); K, A/mallard/Ohio/184/86 (H5N1); L, A/mallard/Minnesota/199106/99 (H3N8); M, A/mallard/Minnesota/355779/00 (H5N2); and N, A/mallard/Minnesota/182761/98 (H7N3).
Sample sizes for HPAI and LPAI virus treatment groups refer to the numbers of birds that met our criteria for confirmed-infected birds. Confirmed-infection status was based on the detection of oropharyngeal viral shedding on or after 2 dpi or fecal shedding at any time. Sample sizes for the sham treatment groups refer to the number of sham-inoculated birds for each species.
bELISA positive based on the manufacturer's diagnostic cutoff of an S/N value of <0.50.
Serologic assay.
Serologic testing was performed on all samples via the AGID test and a commercially available bELISA (FlockCheck AI MultiS-Screen antibody test kit; Idexx Laboratories, Westbrook, ME). The AGID test was performed using published standard procedures (22) and reagents obtained from the National Veterinary Service Laboratory, Ames, IA. The bELISA utilizes a mouse-derived monoclonal antibody against the type A influenza virus NP and was performed and interpreted following procedures described by the manufacturer. Briefly, serum samples were diluted 1:10 in a test sample diluent, and 100 μl of the diluted sample was dispensed into the antigen-coated test plate. After the samples were allowed to incubate for 60 min at room temperature, each well of the plate was washed five times with a test kit wash solution, and 100 μl of anti-AI virus-horseradish peroxidase conjugate was added to each well. The conjugate was allowed to incubate for 30 min at room temperature. Each well of the plate was again washed five times, and 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate solution was dispensed into each well. After a 15-min incubation at room temperature, the reaction was stopped by adding 100 μl of stop solution, and absorbance values were measured and recorded at A650 with a Bio-Rad Benchmark microplate reader (Hercules, CA). Positive- and negative-control samples provided with the test kit were included on each plate. Serum samples with a test sample result-to-negative-control (S/N) absorbance ratio greater than or equal to 0.50 were considered negative for the presence of AI virus antibodies, and samples with an S/N value less than 0.50 were considered positive.
Data analysis.
Sensitivity and specificity were calculated for both the bELISA and the AGID test, along with exact 95% confidence intervals (95% CI). Specificities for the bELISA and the AGID test were calculated based on postinoculation test results for confirmed-uninfected, sham-inoculated birds. Percent agreement and kappa statistics (κ) with asymptotic 95% CI were used to estimate the agreement between tests. Interpretation of the kappa value was based on the Landis and Koch classification (10), where <0.2 indicates a slight agreement, 0.2 to 0.4 indicates a fair agreement, 0.4 to 0.6 indicates a moderate agreement, 0.6 to 0.8 indicates a substantial agreement, and >0.8 indicates an almost perfect agreement.
McNemar's χ2 test was used to determine whether there was a significant difference between the sensitivities of the bELISA and the AGID test performed on paired samples, and the Pearson χ2 test was used to compare sensitivities for birds infected with HPAI virus and birds infected with LPAI virus. One-way analysis of variance was used to compare the S/N ratios for uninfected birds, HPAI virus-infected birds, and LPAI virus-infected birds. Multiple comparisons following a significant overall analysis of variance were performed using the Bonferroni correction to limit the overall type I error rate to 5%. All tests were performed assuming a two-sided alternative hypothesis, and P values of <0.05 were considered statistically significant.
The characteristics of the bELISA were further evaluated using a nonparametric receiver operating characteristic (ROC) curve analysis, in which the sensitivity was plotted against 1 minus the specificity over the entire range of possible cutoff values. The diagnostic cutoff for the bELISA was evaluated in relation to sensitivity, specificity, and cut point-specific likelihood ratio values. All analyses were performed using commercially available statistical software (Stata 10.1; StataCorp, College Station, TX).
RESULTS
The sensitivities, specificities, and 95% CI for the AGID test and the bELISA are summarized in Table 2. The sensitivity of the bELISA was higher than that of the AGID test regardless of whether birds were infected with an HPAI (McNemar's χ2; P = 0.005) or an LPAI (P = 0.001) virus, although both tests had a higher sensitivity with the HPAI virus-infected birds than with the LPAI virus-infected birds (Pearson χ2; P = 0.001 and P = 0.005 for the bELISA and the AGID test, respectively). Neither the bELISA nor the AGID test produced any false-positive results with the 103 confirmed-uninfected birds, both yielding identical specificity estimates of 1.00 (95% CI, 0.965 to 1.00).
TABLE 2.
Sensitivities of the bELISA and the AGID test for serum samples from birds that were inoculated with LPAI and HPAI virusesa
| Virus type | Sample size | AGID test sensitivity (95% CI) | bELISA sensitivity (95% CI) |
|---|---|---|---|
| HPAI | 56 | 0.821 (0.696-0.911) | 0.964 (0.877-0.996) |
| LPAI | 122 | 0.607 (0.514-0.694) | 0.754 (0.668-0.828) |
| Total | 178 | 0.674 (0.600-0.742) | 0.820 (0.756-0.874) |
The specificity for serum samples from 103 sham-inoculated (negative-control) birds was 1.000 (95% CI, 0.965 to 1.000) for both tests.
The measures of agreement between the bELISA and the AGID test are summarized in Table 3. Following the interpretation of kappa proposed by Landis and Koch (10), there was substantial agreement between the tests in the overall populations of infected and uninfected birds, while agreement between the tests for the infected birds ranged from fair (for HPAI virus) to moderate (for LPAI virus). Although the point estimate for kappa for HPAI virus-infected birds was lower than that for LPAI virus-infected birds, there was substantial overlap between the 95% CI, suggesting that this difference was not significant.
TABLE 3.
Measures of agreement between the AGID test and the bELISA for the overall population of birds evaluated and for different subpopulations of infected birds
| Bird population | No. of birds | % Agreement | Kappa (95% CI) |
|---|---|---|---|
| Infected and uninfected | 281 | 86.5 | 0.73 (0.62-0.85) |
| Infected (either pathotype) | 178 | 78.7 | 0.45 (0.31-0.59) |
| HPAI virus infected only | 56 | 85.7 | 0.29 (0.11-0.48) |
| LPAI virus infected only | 122 | 75.4 | 0.45 (0.28-0.62) |
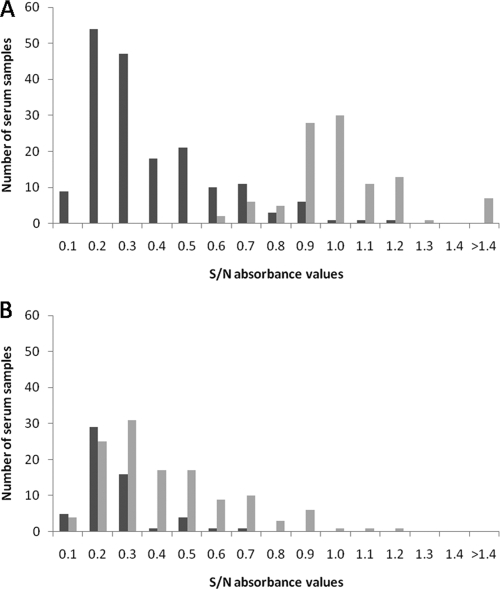
The mean (standard error) bELISA S/N absorbance ratio for the uninfected birds was 0.98 (0.03), while that for the LPAI virus-infected birds was 0.38 (0.02) and that for the HPAI virus-infected birds was 0.21 (0.02). The mean S/N ratio for the uninfected birds was significantly higher than that for either the LPAI virus-infected birds or the HPAI virus-infected birds (P < 0.001), and the mean for the LPAI virus-infected birds was also significantly higher than that for the HPAI virus-infected birds (P < 0.001). The distribution of S/N absorbance ratios from confirmed-infected and -uninfected birds was bimodal, although some overlap between the ranges was apparent (Fig. 1A). The S/N absorbance ratios for serum samples from HPAI virus- and LPAI virus-infected birds overlapped substantially (Fig. 1B).
FIG. 1.
(A) Distribution of S/N absorbance ratios from the bELISA for serum samples from wild avian species inoculated with an AI virus (n = 178) (black) or a sham inoculum (n = 103) (gray). (B) Distribution of S/N absorbance values from the bELISA for birds infected with an LPAI virus (n = 122) (gray) or an HPAI virus (n = 56) (black).
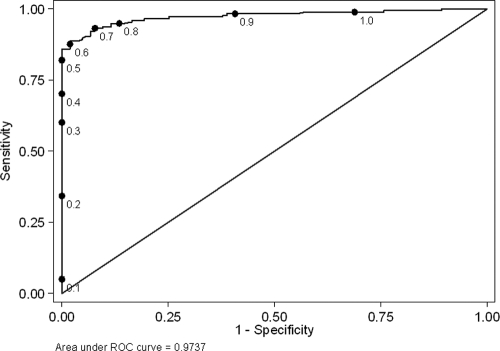
An ROC curve for the bELISA in the populations of 178 infected and 103 uninfected birds is shown in Fig. 2. The area under the ROC curve was 0.974 (95% CI, 0.949 to 0.990), which can be interpreted as the probability that a randomly selected infected bird would have a lower S/N ratio than a randomly selected uninfected bird. While the optimal cutoff point for a diagnostic test is somewhat subjective and dependent on the intended use of the assay, the manufacturer's recommended cutoff (S/N absorbance ratio of 0.50) yielded a specificity estimate of 1.00. Increasing the S/N ratio cutoff above 0.50 increased the sensitivity of the assay but also resulted in a reduction in specificity (Table 4).
FIG. 2.
ROC curve for a bELISA evaluated with a population of 103 uninfected wild birds and 178 wild birds with confirmed AI virus infection. Points on the curve correspond to different cutoff values (S/N ratios) for the test.
TABLE 4.
Characteristics of a bELISA for the diagnosis of AI virus over a range of cutoff valuesa
| Cutoffb (S/N ratio) | Sensitivity (%) | Specificity (%) | Correctly classifiedc (%) | Likelihood ratiod |
|---|---|---|---|---|
| 1.0 | 98.9 | 31.1 | 74.0 | 1.4 |
| 0.9 | 98.3 | 59.2 | 84.0 | 2.4 |
| 0.8 | 94.9 | 86.4 | 91.8 | 7.0 |
| 0.7 | 93.3 | 92.2 | 92.9 | 12 |
| 0.6 | 87.6 | 98.1 | 91.5 | 45 |
| 0.5f | 82.0 | 100 | 88.6 | NCe |
| 0.4 | 70.2 | 100 | 81.1 | NC |
| 0.3 | 60.1 | 100 | 74.7 | NC |
| 0.2 | 34.3 | 100 | 58.4 | NC |
| 0.1 | 5.0 | 100 | 39.9 | NC |
The test population included 103 uninfected wild birds and 178 wild birds with confirmed AI virus infection.
Absorbance of the sample divided by the absorbance of the negative control.
Percentage of birds in the test population with a correctly identified infection status.
The likelihood ratio for a positive test was determined as follows: sensitivity/(1 − specificity).
NC, not calculated, because the estimated specificity at this cutoff was 100%.
Manufacturer's recommended cutoff.
DISCUSSION
As a result of the H5N1 HPAI virus epidemic in Eurasia and Africa, there has been an unprecedented number of resources allocated for AI virus surveillance in wild birds. While most of these efforts have been directed at H5N1 HPAI viruses, there also has been increased interest in improving our understanding of the ecology and natural history of AI viruses in wild birds. To date, agent-based assays, such as PCR and virus isolation, have been the primary diagnostic tools utilized for AI virus surveillance in wild birds. This surveillance approach has been used successfully to identify AI viruses from a wide diversity of free-living avian species (11, 18) and to better define the epidemiology of AI virus in wild-bird reservoirs (19). There are limitations to this approach, however, including cost, the need for a large sample size, and temporal variations in viral shedding that commonly occur in wild birds and result in a limited window of opportunity for AI virus detection. The potential for such seasonal and temporal variations in AI virus infection is often unknown for a given species. Serologic testing for antibodies to AI viruses has the potential to complement existing tools employed for AI virus surveillance in wild birds, particularly when sampling involves avian species, times, or geographic locations for which information on AI virus is lacking. Serologic tests for type A influenza virus antibody detection, however, have traditionally been overlooked as a diagnostic tool for AI virus surveillance in wild birds, likely due to the reported poor sensitivity of the AGID test for waterfowl.
The results of this study indicate that the bELISA examined in this study was a more sensitive serologic test than the AGID test for detecting prior AI virus infection in wild birds. Both the AGID test and the bELISA had excellent specificities, as no false positives were detected among the 103 confirmed-uninfected, sham-inoculated birds that were included in this study. The sensitivities of both assays were higher for HPAI virus-infected birds than for LPAI virus-infected birds, which likely reflects the increased replication of the former, resulting in greater antigen exposure to immunocompetent cells and subsequent humoral response. Although the kappa statistics suggested only fair to moderate agreement between the tests for infected birds, this is explained in part by the downward bias on kappa in populations with a high proportion of positive test results as well as by the difference in test sensitivities (6).
While the performances of the bELISA appeared to vary between wild avian species (Table 1), the low sample size within each species and the differences in experimental design between the studies preclude making precise statements about the accuracy of the bELISA for specific wild avian species. The bELISA, however, was more sensitive than the AGID test for almost every avian species, and this was particularly true for dabbling duck species of the genus Anas. Species in this genus are known important reservoirs for AI virus and consistently have the highest prevalence of AI virus infection among wild avian species. There are limitations on what information the bELISA provides, and positive results should not be overinterpreted. Positive bELISA results provide no information regarding the viral subtype or pathotype or the time when the viral exposure occurred. Based on the results of this study, however, the bELISA does have valid applications for AI virus surveillance in wild birds, both as a preliminary screening test and as a complement to more traditional agent identification methods. As a screening test, the bELISA can provide information on prior exposure to AI virus in avian species that are difficult to sample due to geographic location, habitat utilization, or behavior. Such information could be used to determine whether more costly and time-consuming sampling efforts are warranted. As a complementary diagnostic tool, serologic data provide insight into species that are exposed to AI virus and additional information for interpreting AI virus infection in a given species. While the bELISA did require a plate reader, the test kits themselves were relatively inexpensive and are conducive for high-throughput diagnostic screening, especially if performed with an automated plate washer. Finally, serologic testing provides a diagnostic tool and surveillance option that does not require working with infectious virus. As such, serology provides added benefits for lab safety and ease of shipping diagnostic samples.
It is important to note that the current study was performed with serum samples collected from birds experimentally inoculated 10 to 21 dpi and therefore is likely to yield higher sensitivity estimates than would be encountered when the test is applied to field samples. The immune responses vary between different species of birds (7), and for many avian taxa, these responses are poorly understood. Consequently, additional studies are warranted to evaluate the sensitivity of the bELISA with experimental studies of longer duration and/or with field samples in order to further validate this serologic assay.
Acknowledgments
Funding for this work was provided by ARS CRIS project 6612-32000-048-00D and Specific Cooperative Agreement 58-6612-2-0220 between the Southeast Poultry Research Laboratory and the Southeastern Cooperative Wildlife Disease Study. Additionally, this work was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Department of Health and Human Services, under contract no. HHSN266200700007C.
The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
We thank Joan R. Beck for excellent technical assistance.
Footnotes
Published ahead of print on 22 April 2009.
REFERENCES
- 1.Brown, J. D., D. E. Stallknecht, J. R. Beck, D. L. Suarez, and D. E. Swayne. 2006. The susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg. Infect. Dis. 121663-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, J. D., D. E. Stallknecht, S. Valeika, and D. E. Swayne. 2007. Susceptibility of wood ducks to H5N1 highly pathogenic avian influenza virus. J. Wildl. Dis. 43660-667. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J. D., D. E. Stallknecht, and D. E. Swayne. 2008a. Experimental infection of swans and geese with highly pathogenic avian influenza virus (H5N1) of Asian lineage. Emerg. Infect. Dis. 14136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, J. D., D. E. Stallknecht, and D. E. Swayne. 2008b. Transmission of H5N1 high pathogenicity avian influenza virus to herring gulls (Larus argentatus) through intranasal inoculation of virus and ingestion of virus-infected chicken meat. Avian Pathol. 37393-397. [DOI] [PubMed] [Google Scholar]
- 5.Cattoli, G., and I. Capua. 2007. Diagnosing avian influenza in the framework of wildlife surveillance efforts and environmental samples. J. Wildl. Dis. 43(Supplement)S35-S39. [Google Scholar]
- 6.Feinstein, A. R., and D. V. Cicchetti. 1990. High agreement but low kappa. I. The problems of two paradoxes. J. Clin. Epidemiol. 43543-549. [DOI] [PubMed] [Google Scholar]
- 7.Higgins, D. A. 1996. Comparative immunology of avian species, p. 149-205. In T. F. Davison, T. R. Morris, and L. N. Payne (ed.), Poultry immunology, 24th ed. Carfax Publishing Co., Abingdon, United Kingdom.
- 8.Hinshaw, V. S., R. G. Webster, and B. Turner. 1980. The perpetuation of orthomyxoviruses and paramyxoviruses in Canadian waterfowl. Can. J. Microbiol. 26622-629. [DOI] [PubMed] [Google Scholar]
- 9.Hinshaw, V. S., G. M. Air, A. J. Gibbs, L. Graves, B. Prescott, and D. Karunakaran. 1982. Antigenic and genetic characterization of a novel hemagglutinin subtype of influenza A viruses from gulls. J. Virol. 42865-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landis, J. R., and G. G. Koch. 1977. The measurement of observer agreement for categorical data. Biometrics 33159-174. [PubMed] [Google Scholar]
- 11.Olsen, B., V. J. Munster, A. Wallensten, J. Waldenstrom, A. D. M. E. Osterhaus, and R. A. M. Fouchier. 2006. Global patterns of influenza A virus in wild birds. Science 312384-388. [DOI] [PubMed] [Google Scholar]
- 12.Pantin-Jackwood, M. J., D. L. Suarez, E. Spackman, and D. E. Swayne. 2007. Age at infection affects the pathogenicity of Asian highly pathogenic avian influenza H5N1 viruses in ducks. Virus Res. 130151-161. [DOI] [PubMed] [Google Scholar]
- 13.Perkins, L. E. L., and D. E. Swayne. 2001. Pathobiology of A/chicken/Hong Kong/220/97 (H5N1) avian influenza virus in seven gallinaceous species. Vet. Pathol. 38149-164. [DOI] [PubMed] [Google Scholar]
- 14.Perkins, L. E. L., and D. E. Swayne. 2003. Varied pathogenicity of a Hong Kong-origin H5N1 avian influenza virus in four passerine species and budgerigars. Vet. Pathol. 4014-24. [DOI] [PubMed] [Google Scholar]
- 15.Slemons, R. D., and B. C. Easterday. 1972. Host response differences among 5 avian species to an influenzavirus-A/turkey/Ontario/7732/66 (Hav5N?). Bull. W. H. O. 47521-525. [PMC free article] [PubMed] [Google Scholar]
- 16.Snyder, D. B., W. W. Marquardt, F. S. Yancey, and P. K. Savage. 1985. An enzyme-linked immunosorbent assay for the detection of antibody against avian influenza virus. Avian Dis. 29136-144. [PubMed] [Google Scholar]
- 17.Spackman, E., D. L. Suarez, and D. A. Senne. 2008. Avian influenza diagnostics and surveillance methods, p. 299-308. In D. E. Swayne (ed.), Avian influenza. Blackwell Publishing, Ames, IA.
- 18.Stallknecht, D. E., and S. M. Shane. 1988. Host range of avian influenza virus in free-living birds. Vet. Res. Commun. 12125-141. [DOI] [PubMed] [Google Scholar]
- 19.Stallknecht, D. E., and J. D. Brown. 2007. Wild birds and the epidemiology of avian influenza. J. Wildl. Dis. 43(Supplement)S15-S20. [Google Scholar]
- 20.Suarez, D. L., and S. Schultz-Cherry. 2000. Immunology of avian influenza virus: a review. Dev. Comp. Immunol. 24269-283. [DOI] [PubMed] [Google Scholar]
- 21.Swayne, D. E., and D. A. Halvorson. 2008. Influenza, p. 153-184. In Y. M. Saif, A. M. Fadly, J. R. Glisson, L. R. McDougald, L. Nolan, and D. E. Swayne (ed.), Diseases of poultry, 12th ed. Blackwell Publishing, Ames, IA.
- 22.Swayne, D. E., D. A. Senne, and D. L. Suarez. 2008. Influenza, p. 128-134. In L. Dufour-Zavala, D. E. Swayne, J. R. Glisson, J. E. Pearson, W. M. Reed, M. W. Jackwood, and P. R. Woolcock (ed.), Isolation and identification of avian pathogens, 5th ed. American Association of Avian Pathologists, Athens, GA.
- 23.Swayne, D. E., and R. D. Slemons. 2008. Using mean infectious dose of high- and low-pathogenicity avian influenza viruses originating from wild duck and poultry as one measure of infectivity and adaptation to poultry. Avian Dis. 52455-460. [DOI] [PubMed] [Google Scholar]