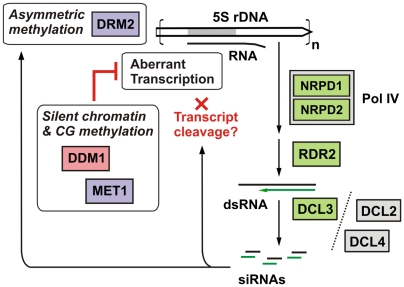
Figure 6. Model for 5S rDNA methylation and aberrant transcript silencing.
A canonical RNA polymerase (e.g., Pol III) is assumed to initiate at the 5S rRNA gene promoter, transcribe through the 5S rRNA gene (120 bp, gray), and continue into the intergenic spacer (IGS, white). Concurrently, dsRNA corresponding to the IGS is generated in a Pol IV and RDR2-dependent manner. DCL3 processes dsRNA into 24-nt siRNAs, with DCL2 and DCL4 being alternate enzymes at this step. RNA-directed DNA methylation (RdDM) resulting from this process is thought to require DRM2. DDM1/MET1 maintenance of silent chromatin and CG methylation represses aberrant IGS transcription, limiting production of siRNA precursors and, by consequence, attenuating RdDM. IGS-derived siRNAs may also guide cleavage of nascent IGS transcripts.