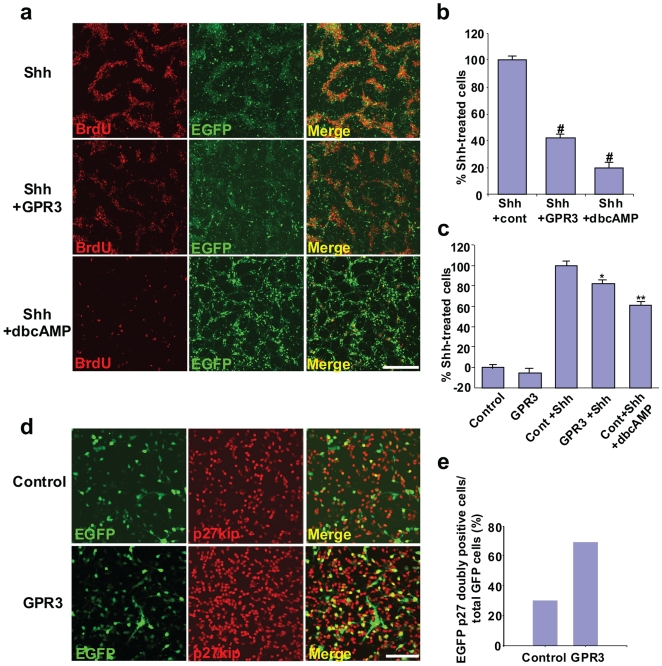
Figure 1. Exogenous GPR3 partially inhibits Shh-induced proliferation of rodent cerebellar granule neurons and is associated with increased p27/Kip expression.
GCPs isolated from rat cerebellum (P7) were electroporated with a GPR3/EGFP or control vector using the AMAXA nucleofector system. Shh (1 ug/ml) was then added 6 hours later. Thirty hours later, BrdU (final concentration = 10 µM) was added 12 hours prior to fixation. Cell proliferation was determined using BrdU incorporation assessed by immunohistochemistry with a BrdU-specific monoclonal antibody (panels a, b). dbcAMP (100 µM) was used as a chemical cAMP activator and as a positive control. In panel a, representative fields from dishes are visualized from the Shh and control vector group (top row), Shh and GPR3/EGFP-electroporated group (middle row) and the Shh, control vector group and dbcAMP (bottom row). Visualization by BrdU immunohistochemistry (red dye), EGFP fluorescence (green dye), and the combined merged image are shown. Five random fields were selected per each dish (n = 3) from the Shh, Shh+GPR3 and Shh+GPR3+cAMP groups. The number of GFP-positive cells (representing positively transfected neurons) was counted (at least one hundred GFP-positive neurons per each field). Doubly labeled cells (yellow color) were also enumerated per each dish. Values were then expressed as the percentage of doubly-labeled cells out of total GFP positive cells in the Shh+GPR3 group or Shh+GPR3+dbcAMP groups compared to Shh group, i.e. % Shh-treated cells (panel b). In parallel, cell proliferation was also determined using an ELISA assay based on the measurement of BrdU incorporation during DNA synthesis (Panel C) Variations between percentage of cells on the plates in panel b and c are likely due to differences in assays (visual counting vs. counting by colorimetry). For p27/kip1 expression (panel d, e), GCPs isolated from rat cerebellum (P4) were electroporated with the GPR3/EGFP expression (bottom row, panel d) or control vector (top row, panel d). p27/kip1 expression was detected by immunohistochemistry, 48 hours later. EGFP and p27/kip1 doubly positive cells were counted in GPR3 and control groups (panel e). *p<0.01;**p<0.001, # p<0.0001, scale bar = 200 µm.