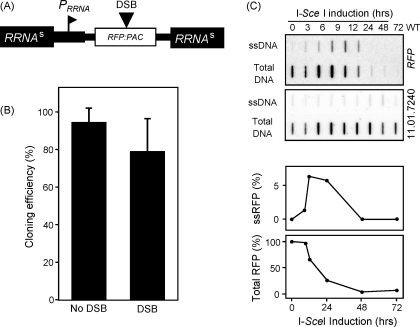
Fig. 1.
Response to a DSB at the RRNA-spacer locus. (A) The schematic map illustrates an RFP–PAC fusion gene (RsP) with an embedded I-SceI site (indicated by DSB) at the RRNA-spacer (RRNAs) locus. pRsPRRNA was assembled as follows: an RRNA promoter (PRRNA) fragment was amplified from genomic DNA using primers RpF (GATCcggcggTAGCTTTCCACCCAGCGC) and RpR (GATCcggccgggcccACTGggatccTCTGAGAGCGGTCAGTTGC), digested with EagI (relevant restriction sites in lower-case) and ligated to a NotI-digested RRNA-spacer fragment in pBlusescript. An RsP cassette was then added using the BspI201 and BamHI sites. The RsPRRNA construct was then digested with SacI/AgeI and introduced into the 2T1 bloodstream-form T. brucei strain [10] that also contained a tetracycline-inducible I-SceI ORF introduced using the pRPai construct [17]. These Lister 427, clone 221a cells were grown and manipulated as described [10]. (B) A clonogenic assay to assess recovery from a DSB. Cells in all un-induced wells tested remained puromycin-resistant and cells in every induced well were puromycin-sensitive indicating loss of the RsP cassette in the latter case. Cell counts were carried out using a haemocytometer and tetracycline (used at 1 μg ml−1) was from Sigma. Data are derived from a pair of independent RsPRRNA strains and error bars represent one standard deviation. (C) Physical monitoring of DNA resection adjacent to the lesion was carried out by slot-blot assay as described [1]. Genomic DNA samples were ‘native’, to detect ssDNA or denatured, to detect total DNA. The probes used on each blot are indicated on the right; the control probe is from chromosome 11 (Tb11.01.7240). Phoshorimager analysis was used to quantify the signals and ssRFP values were derived after correction for background, ssDNA versus total DNA and loading. The RFP ssDNA and total DNA plots indicate resection kinetics and DNA loss respectively.