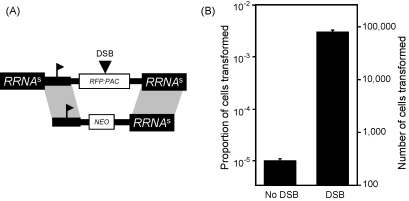
Fig. 2.
A genomic DSB increases transformation efficiency. (A) The schematic map illustrates the genomic target (reproduced from Fig. 1A) and the exogenous DNA construct. (B) Transformation assays. A DSB was induced by growth in tetracycline (1 μg ml−1) for 3 h. Nucleofection (Lonza) was carried out as described [7]. Briefly, 2.5 × 107 RsPRRNA cells were resuspended in 100 μl of human T-cell Nucleofector solution, mixed with 10 μg of purified linear DNA and subjected to Nucleofection using program X-001 in a 1 mm-gap cuvette. G418 was added <6 h later at 2 μg ml−1. To estimate the number of transformed clones, we initially used serial dilutions in 96-well plates but, due to concerns with loss of accuracy during extensive serial dilution, we used a modified approach to generate the data presented. Briefly, in duplicate experiments, 6 h after Nucleofection and drug addition, we distributed a sample predicted to contain 32 transformants (based on estimates from serial dilutions) over a 96-well plate. This approach yielded 15–40% positive wells per plate and was therefore deemed to have provided accurate scores. Error bars represent one standard deviation.