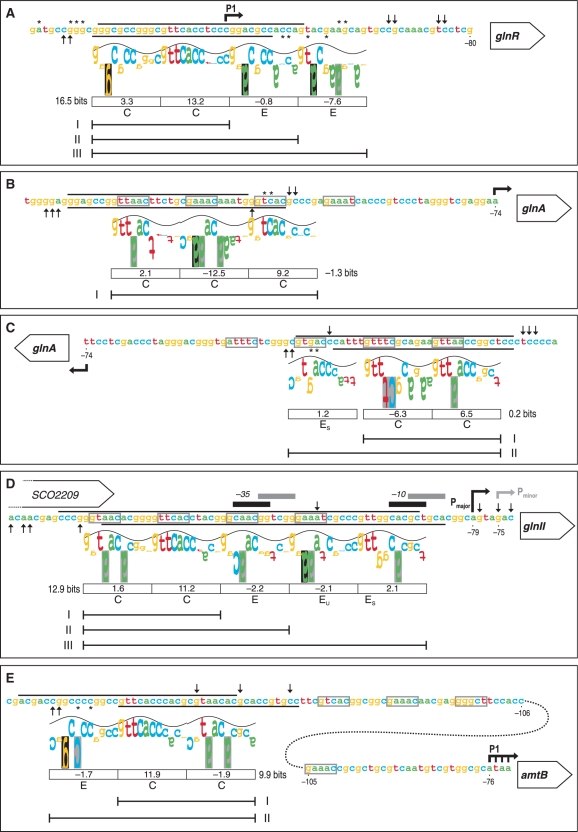
Figure 3.
‘Information content’ analysis of the new PhoP-binding sites identified in this work using the sequence walker method (29). The footprinting results of the promoter regions of glnR (A), glnA (B, C), glnII (D) and amtB (E) are summarized on the nucleotide sequences. The protected regions of the upper and bottom strands are indicated by the respective lines. Symbols for partially protected (asterisk) and hypersensitive sites (upward arrow, downward arrow) are as in Figure 2. For the PhoP-binding site in glnA, the two possible walkers are shown in the coding (B) and complementary strands (C). The walker limits are 2 bits, which is also the top of the sine wave, and −3 bits at the bottom. The height of each letter is the individual information value, determined from the Model I weight matrix (11). Negative values are represented by upside down letters. When a base does not occur at a given position in the set of sequences which forms the model, the letter background is black; a grey background indicates that the letter extends beyond the lower limit (−3 bits). The sine wave has a periodicity of 10.6 bases (the helical twist of B-form DNA) and serves to indicate major/minor groove contacts (28). The following features are bellow the walkers: boxes containing the Ri values for the above 11 nt direct repeat unit (DRu), the total Ri for the core site, letter codes for the DRu structure [C for core, E for extension, EU for extension unstable, ES for extension support (11)], and line segments comprising the DRus bound by PhoP monomers in the retarded complexes observed in Figure 1. The conserved 5-mer sequences of the GlnR-binding sites are boxed (grey lines). Bent arrows are the transcription start points. The −35 and −10 elements of major and minor glnII promoters are also shown.