Abstract
Type I restriction endonucleases are intriguing, multifunctional complexes that restrict DNA randomly, at sites distant from the target sequence. Restriction at distant sites is facilitated by ATP hydrolysis-dependent, translocation of double-stranded DNA towards the stationary enzyme bound at the recognition sequence. Following restriction, the enzymes are thought to remain associated with the DNA at the target site, hydrolyzing copious amounts of ATP. As a result, for the past 35 years type I restriction endonucleases could only be loosely classified as enzymes since they functioned stoichiometrically relative to DNA. To further understand enzyme mechanism, a detailed analysis of DNA cleavage by the EcoR124I holoenzyme was done. We demonstrate for the first time that type I restriction endonucleases are not stoichiometric but are instead catalytic with respect to DNA. Further, the mechanism involves formation of a dimer of holoenzymes, with each monomer bound to a target sequence and, following cleavage, each dissociates in an intact form to bind and restrict subsequent DNA molecules. Therefore, type I restriction endonucleases, like their type II counterparts, are true enzymes. The conclusion that type I restriction enzymes are catalytic relative to DNA has important implications for the in vivo function of these previously enigmatic enzymes.
INTRODUCTION
Restriction–modification (R–M) systems are part of the global mechanism designed to maintain the integrity of bacterial genomes (1,2). These systems have dual functions as they serve to protect the host genome against restriction by methylating residues within target sequences, while simultaneously functioning to destroy foreign DNA. As these enzymes confer the ability to distinguish self from non-self, they can be viewed as part of a rudimentary but effective bacterial immune system (3).
R–M enzymes are classified into three main groups designated types I–III, based on their subunit structure, cofactor requirements, sequence recognition and cleavage position (4–6). Type II systems are the most well known, having found widespread use in molecular cloning. They comprise separate methylases and homodimeric restriction endonucleases which act independently from one another and which methylate or cleave a single, 4–8 bp palindromic DNA sequence. Cleavage is catalytic and typically occurs either within, or immediately adjacent to the palindromic sequence (7). Type III R–M enzymes are tetrameric holoenzymes that possess sequence-specific methylation, restriction and DNA dependent nucleoside triphosphatase activities (5). The sequence recognized is 5–6 bp in length and cleavage typically occurs 25–27 bp away from, and to one side of the recognition sequence.
The most complex of the four groups are the type I enzymes which were the first R-M systems discovered (1). They have since been discovered in a number of Eubacteria and the Archaeabacteria as well (6). Type I restriction endonucleases (REs) consist of three different subunits: methylase (M), restriction (R) and specificity (S) encoded by the hsdM, -R and -S genes, where hsd = Host Specificity for DNA (1). Together, they form an intriguing, multifunctional complex which can either restrict or modify DNA. Here, the mode of action of the complex is dictated by the methylation state of the recognition sequence (8). A fully methylated site results in no action being taken and in enzyme dissociation; hemi-methylated target sequences direct the enzyme into a protective methylation mode producing fully methylated DNA, while unmethylated DNA shifts the enzyme into a destructive (and protective) restriction mode. It is in this protective mode that type I enzymes restrict foreign DNA and thereby maintain the integrity of the host genome.
Type I REs are divided into four families designated IA–ID, based on amino acid conservation, antibody cross-reactivity and enzymatic properties (1,5). The prototype of family IC is the plasmid-encoded EcoR124I. This enzyme is unique in its cofactor requirements as S-adenosyl methionine (SAM) is non-essential to the ATPase and restriction endonuclease activities (9). Similar to other Type I REs, EcoR124I holoenzyme is a pentameric complex of three subunits, with a stoichiometry of R2:M2:S1 (10,11). HsdS contains two sequence recognition domains that impart sequence specificity to both the methylation and restriction activities of the holoenzyme. The HsdM subunits methylate adenine bases within the target sequence (GAAn6RTCG) using SAM as a donor. The methytransferase (MTase; M2:S1) is sufficient for DNA methylation, acting in catalytic fashion relative to DNA (12). The HsdR subunits contain the Helicase SuperFamily 2 motifs and endonuclease active site which impart translocation and restriction activities to the holoenzyme, respectively (1,13).
To restrict DNA, these ∼400 kDa complexes first recognize and bind to specific, non-methylated, bi-partite and asymmetric DNA sequences embedded within double-stranded DNA (dsDNA) (14). These sequences consist of two specific domains (3–5 bp in length) split by a non-specific spacer of defined length (6–8 bp) (2). Once bound to the target sequence, the enzyme hydrolyzes ATP with a kcat of ∼100 000 min−1, while simultaneously translocating dsDNA bidirectionally toward the holoenzyme anchored at the recognition sequence (11,14,15). When translocation is impeded, typically thousands of base pairs distant from the recognition sequence, restriction occurs (14,16–18). Double-stranded DNA cleavage involves sequential nicking of each strand of the duplex in close proximity by diametrically opposed R-subunits (17,19,20). Surprisingly, these enzymes are thought to remain bound to the nascent cleaved DNA where they continue to rapidly hydrolyze ATP for several hours (11,21–23). Thus, they have been viewed as catalytic relative to ATP but stoichiometric relative to DNA (8,22,23). That is, each holoenzyme is able to cleave only a single DNA substrate. This facet of enzyme function has not been rigorously addressed to date.
The failure to mediate multiple rounds of DNA cleavage represents an unusual state of affairs within a cell as this would result in a series of ∼5 kB DNA fragments with a type I RE still bound turning over substantial amounts of ATP with a kcat of 100 000 min−1 (11). This would require any one of a number of mechanisms to neutralize these complexes including the action of nucleases, proteases or combinations thereof (1,11). Failure to neutralize these complexes could potentially deplete the intracellular ATP pools and ensure the death of a phage-infected cell (24). Although this would be detrimental to the cell in question, the remaining uninfected cells in the population would survive. Since the role of the R–M system is to protect the host, the persistence of these enzyme–DNA complexes could be viewed as suicidal. However, restriction proficient hosts do survive infection by unmodified phages (1). This suggests that efficient mechanisms are in place to remove DNA-bound, post-cleavage enzyme or conceivably that the enzyme may frequently dissociate into its component parts as a means to control enzyme function (25,26). Alternatively, Type I REs may release the DNA following cleavage and act in catalytic fashion relative to DNA.
To more clearly understand the type I RE cleavage mechanism, the study of DNA cleavage by EcoR124I was undertaken. To our surprise, we discovered that contrary to the view held for over 35 years, type I RE are not stoichiometric but are instead catalytic with respect to DNA. Further, the mechanism involves formation of a dimer of holoenzymes, with each monomer bound to a target sequence and, following cleavage, each dissociates in an intact form to bind and restrict subsequent DNA molecules. The fact that type I restriction enzymes are catalytic has broad implications to the in vivo biology of the R–M enzyme family.
MATERIALS AND METHODS
Chemicals
All chemicals were as described previously (11). ATP (GE Biosciences) was dissolved in 1 M Tris–HCl (pH 8.0) and stored as aliquots at −20°C. ATPγS (Roche) and ADP (Sigma-Aldrich) were dissolved in 0.5 M Tris–HCl (pH 8.0) just before use. The concentrations of ATP, ATPγS and ADP were determined spectrophotometrically at 260 nm using ϵ = 15 400 M−1 cm−1.
Purification of the EcoR124I holoenzyme
EcoR124I was purified exactly as described (11). The concentration of holoenzyme was determined at 280 nm using ϵ = 366 090 M−1 cm−1 (27).
Plasmid DNA
The plasmids used in this study are pDRM.1R (R+; 2891 bp), pDRM.2R (2R+; 2921 bp), pPB67 (R0; 4363 bp), pPB248 (R+; 4266 bp) and pPB455 (R+; 9826 bp). Negatively supercoiled DNA was purified as described previously (11). The nucleotide concentration of DNA was determined at 260 nm using ϵ = 6500 M−1 cm−1. The concentration of DNA in all experiments is reported in nanomolar molecules.
ATPase assay
Hydrolysis of ATP was monitored using a continuous spectrophotometric ATPase assay (11). Reactions were done at 25°C and contained 20 mM Tris–acetate (pH 7.5), 100 μg/ml of BSA, 5% glycerol, 1 mM DTT, 1 mM ATP, 5 mM Mg(OAc)2, 20 U/ml of pyruvate kinase, 20 U/ml of lactate dehydrogenase (LDH), 7.5 mM phosphoenolpyruvate, 0.3 mM NADH, 5nM (-)scDNA (R+; pDRM.1R or 2R+; pDRM.2R) and EcoR124I as indicated. The analysis of data was exactly as described (11).
DNA cleavage assays
Agarose gel-based assays were used to monitor cleavage of (-)scDNA by EcoR124I. Reaction conditions (300 μl) were the same as those used for the ATPase assay except LDH and NADH were omitted. Assays were done either on ice (0°C), or at 25 or 37°C, producing identical results but at rates corresponding to the reaction temperature (data not shown). Following DNA cleavage, time points were stopped by the addition of ficoll gel loading dye (28) containing 0.1% SDS, 10 mM EDTA and 0.4 mg/ml proteinase K (final concentrations). Samples were incubated at room temperature until the last time point was collected, loaded onto a 0.8% agarose gel and subjected to electrophoresis in TAE buffer (40 mM Tris–acetate and 2 mM EDTA) at 700 V. h. Gels were stained with ethidium bromide (0.5 μg/ml in dH20) for 1 h and subsequently destained in dH20 for 20 min. Destained gels were photographed using a DIGI-DOC gel documentation system (UVP, Inc.) and analyzed using ImageQuant v 5.0 software (Molecular Dynamics). To determine the rate of DNA cleavage, the amount of (-)scDNA present in each lane was determined using the volume quantitation function of ImageQuant and expressed as a fraction of the input amount of DNA substrate. In reactions where the amount of linear dsDNA was quantitated, the amount of DNA present is expressed as a fraction of the total dsDNA present in that lane. For other reactions, the method of quantitation is indicated in the figure legend.
Denaturing gel filtration
1.2 mg of EcoR124I was mixed with buffer A [50 mM Tris–HCl (pH 7.5) and 2 mM DTT] containing 0.5 to 6 M guanidine hydrochloride (GuHCl) in separate experiments. The protein sample (1 ml) was applied to a Superose 6 column (24 ml; Amersham Biosciences) equilibrated in the buffer A containing the same concentrations of GuHCl. Following chromatography, fractions were dialyzed against buffer A overnight and subjected to electrophoresis in SDS–PAGE gels followed by gel analysis to evaluate composition. Gels were stained with either Coomassie brilliant blue or silver (SilverSnap II; PIERCE) and analyzed using ImageQuant v5.0 (Molecular Dynamics).
Native gel filtration
EcoR124I–DNA complexes (550 µl; 38–40 nM DNA, 62–70 nM active enzyme in separate reactions), were applied to the Superose 6 column equilibrated in buffer B [20 mM Tris–HCl (pH7.5), 1 mM DTT, 5 mM Mg(OAc)2 and 150 mM NaCl]. Thereafter, DNA–protein complexes were subjected to electrophoresis in agarose gels to ascertain DNA content and cleavage state. Similarly, fractions from the complex and free protein peaks were subjected to electrophoresis in SDS–PAGE gels followed by analysis using ImageQuant to ascertain protein content and assembly state. Separately, the DNA protein complex was added to ATPase and DNA cleavage assays.
To determine enzyme assembly state at the mid-point, the reaction mixture (identical to cleavage assays) was prepared on ice (0°C) and initiated by adding ATP. After 2 min, GuHCl was added to a final concentration of 1 M. The reaction mixture was then applied to the column equilibrated in buffer B. For the end-point, reactions were assembled as above and at t = 30 min, the entire reaction was applied to the column. Following chromatography, fractions were dialyzed and subjected to electrophoresis separately in agarose and SDS–PAGE gels followed by analysis.
The mid- and end-points of the reaction were also subjected to denaturing gel filtration using buffer A containing either 0.5 or 1 M GuHCl. Following chromatography, peak fractions were dialyzed against buffer A and subjected to electrophoresis in both agarose and SDS–PAGE gels. Positions of migration of all bands were compared to a molecular weight marker present in all gels (data not shown).
RESULTS
Definition of terminology used
Before presenting our data and to make the presentation clear, several definitions are necessary. For DNA substrates, we will use the designation R0 to indicate DNA without the target sequence GAAn6RTCG; R+ to indicate a substrate with one sequence and 2R+ to indicate DNA containing two sequences. We define the EcoR124I holoenzyme as a five subunit or pentameric complex with a subunit stoichiometry of R2M2S1. It follows that a dimer of holoenzymes is defined as two pentameric complexes or two, R2M2S1 complexes. The EcoR124I protein used in all experiments is purified as the intact holoenzyme with a subunit stoichiometry of R2M2S1 as demonstrated previously (11). The total concentration of purified protein is measured spectrophotometrically at 280 nm and calculated using the extinction coefficient as described in the Materials and Methods section. The fraction of total protein which is active is termed the ‘active fraction of enzyme’ and is obtained from the saturation point of ATPase assays using R+-DNA as described previously (11). In this work, the level of active enzyme constitutes 22% of the total protein. The reason for the low level of activity is unclear, but is addressed in the last section of the Results.
Is EcoR124I catalytic with respect to DNA?
For the past 35 years, type I RE have been thought of as stoichiometric enzymes. This title has been ascribed to these proteins because they are only capable of performing a single round of DNA cleavage. Subsequent cleavage events, that is multiple rounds of DNA cleavage, were not thought to occur as the protein remained bound to the nascent, restricted DNA molecule where it continued to hydrolyze ATP for several hours (11,21–23). This view has not been rigorously challenged until now.
To determine whether EcoR124I can act in catalytic fashion relative to DNA, a sequential DNA-cleavage assay using two distinct-sized, (-)scDNA substrates was used and the fate of each DNA determined. We reasoned that if EcoR124I is catalytic, then addition of the second (-)scDNA substrate to a completed cleavage reaction should result in cleavage of that DNA as well. In contrast, if the protein is not catalytic, then the second DNA would not be cleaved.
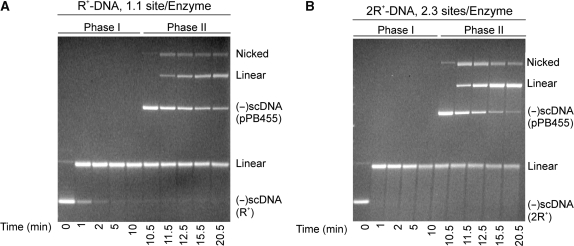
Cleavage assays were done as described in Materials and Methods section and the results are presented in Figure 1. In these experiments, near stoichiometric ratios of active enzyme to DNA molecules were used (4.4–5 nM, respectively). In the first experiment, the DNA substrate present in phase I was negatively supercoiled, R+-DNA 2.9 kB in size and pPB455 (R+-DNA; 9.8 kB) was added as the second DNA at 10 min to initiate phase II. The results show that the first DNA was rapidly and completely cleaved (100% in 5 min) and ∼25% of the second DNA was cleaved in phase II (Figure 1A).
Figure 1.
EcoR124I cleaves two DNA substrates sequentially. Representative agarose gels from sequential cleavage assays are presented. Reactions were done at 37°C and contained 4.4 nM active EcoR124I holoenzyme and 5 nM DNA (2.9 kB) in phase I. To initiate phase II, 5 nM pPB455 (R+-DNA; 9.8 kB) was added at t = 10 min. Following reaction completion, samples were subjected to electrophoresis in agarose gels as described in Materials and Methods section. In (A), the first DNA substrate was R+-DNA and in (B), the first substrate was 2R+-DNA. The position of migration of each band was compared to molecular weight markers present in the same gel (data not shown).
In the second experiment, 2R+-DNA (2.9 kB) replaced R+-DNA in phase I, and pPB455 (9.8 kB; R+-DNA) was added once cleavage of the first DNA was complete. In this experiment the concentration of DNA in phase I is still 5 nM but the concentration of sites is 10 nM, 2.3-fold higher than that of active enzyme (4.4 nM). The resulting agarose gel shows that the 2R+-DNA was rapidly and completely cleaved in <1 min and pPB455 was also efficiently cleaved, proceeding to completion within 10 min following addition (Figure 1B). These results suggest that EcoR124I functions catalytically with respect to DNA as both DNA substrates were cleaved in these reactions. Cleavage of the second DNA is not due to an excess of active enzyme present in the reaction. This follows because the total concentration of sites in phase II of the R+-DNA reaction is 10 nM and in the 2R+-DNA reaction it is 15 nM. In the latter case, the ratio of sites to active enzyme is 3.4 and all of the DNA present was cleaved. We note that at the same concentration of active enzyme, the rate of cleavage of 2R+-DNA is faster than that of R+-DNA. This is an important point and is addressed in the last section of the Results.
Nucleoside cofactors reduce non-specific DNA binding by EcoR124I
As the enzyme preparation is 22% active [determined from the saturation point of ATPase assays using R+-DNA (11)], there is a significant fraction of free protein that could influence the outcome of the reactions presented in the preceding section so as to ‘appear’ catalytic. A model for this was derived from studies using reconstituted enzyme produced by mixing a several-fold excess of R-subunits and methyltransferase together just before use. It proposes that the methyltransferase (M2S1) loads R-subunits onto the DNA and that these same R-subunits dissociate frequently into solution during the course of a reaction. This is followed by reassociation with methytransferase in solution, forming additional ‘active’ enzymes that could cleave subsequent DNA molecules (25,26). Thus the free protein provides a pool with which R-subunits could continuously associate to provide the appearance of catalytic behavior. It is therefore critical to remove unbound protein to more clearly ascertain whether catalytic function can be ascribed to the DNA-bound holoenzyme. To achieve this, we utilized gel filtration and purified stoichiometric complexes of the EcoR124I holoenzyme and R+-DNA.
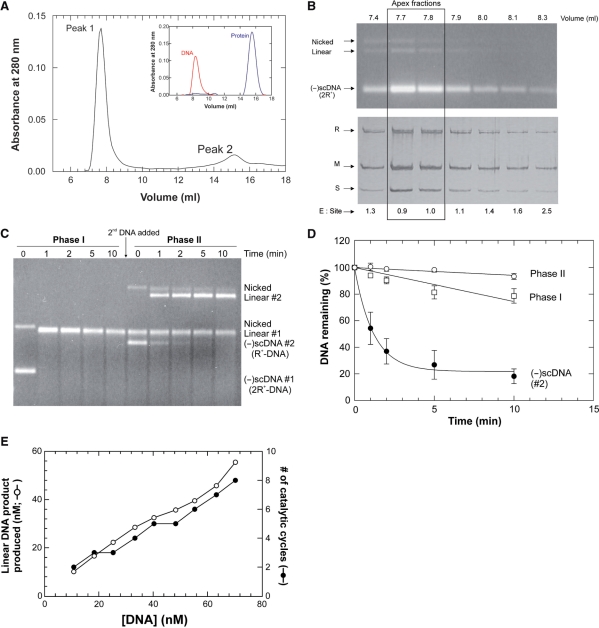
Control gel filtration experiments were done first to determine whether (-)scDNA, protein and the enzyme–DNA complex could be well separated from one another. The results from separate gel filtration runs of DNA and protein only, show that these components elute at 8.3 and 15.8 ml, respectively (Figure 2A, inset). When enzyme–DNA complexes formed using an excess of active enzyme relative to sites were subjected to gel filtration, the resulting profile demonstrated two well separated peaks. The first, larger peak eluted at 7.8 ml, while the second, smaller peak eluted at 15 ml (Figure 2A). Aliquots from resulting fractions from each peak were subjected to agarose and SDS–PAGE gel analysis to determine peak composition. The analysis shows that the first peak contained enzyme and DNA, consistent with the presence of the complex, while the second peak contained free protein only (data not shown).
Figure 2.
The EcoR124I restriction endonuclease is a catalytic enzyme. (A) Gel filtration permits purification of the active EcoR124I–DNA complex. A typical gel filtration elution profile is shown. The binding reaction contained 20 nM molecules of 2R+-DNA and 35 nM active EcoR124I. Peak 1, the enzyme–DNA complex and peak 2, free protein. The composition of each peak was determined using agarose and SDS–PAGE gels. The height of the two peaks varied according to the amount of enzyme relative to DNA in the initial binding reaction (data not shown). Inset: separate, gel filtration traces of (-)scDNA (red) or EcoR124I (blue) only. (B) Analysis of the enzyme–DNA peak following gel filtration reveals a stoichiometric complex. To determine the identity and composition of each fraction in each peak following gel filtration, samples from gel filtration experiments shown in (A), were subjected to electrophoresis in either agarose or SDS–PAGE gels (top and bottom panels, respectively). For this experiment, the binding reaction contained 40 nM sites (2R+-DNA) and 50 nM active enzyme. Each gel was photographed immediately after staining and analyzed using ImageQuant software. In each gel, the amount of DNA or protein present in each fraction was determined by comparison to a titration of either DNA or protein present in the same gel (not shown). These amounts were then used to calculate the ratios of enzyme to sites which are indicated at the bottom of each lane of the SDS–PAGE gel. (C) EcoR124I is catalytic relative to DNA. 2R+–EcoR124I complexes (40 nM sites; 70 nM active enzyme) were purified by gel filtration, aliquots added to reaction mixtures on ice, transferred to 37°C and reactions initiated by addition of ATP to 1 mM, final. At t = 10 min, 5 nM pPB248 [R+; (-)scDNA #2; 4.3 kB] was added and the time course continued. (D) Analysis of the gel shown in (C). Closed circles, pPB248 (-)scDNA present in phase II. Open symbols are the nascent linear dsDNA product produced in phases I and II. The analysis is of three separate reactions from separate chromatography runs done on different days. Quantitation of (-)scDNA was done as described in Materials and Methods section. The amount of linear DNA product present in each lane is expressed as a fraction of the amount present at the 1 min time point of each phase. (E) The EcoR124I enzyme can perform multiple rounds of DNA cleavage. Purified, stoichiometric 2R+–EcoR124I complexes were added to reaction mixtures on ice and increasing amounts of pPB455 ranging from 10 to 69 nM molecules added. The mix was transferred to 37°C and reactions initiated by addition of ATP (1 mM, final) and allowed to proceed to completion. Thereafter samples were subjected to electrophoresis in an agarose gel, stained with ethidium bromide, photographed and analyzed using ImageQuant. The total amount of linear product produced resulting from cleavage of both 2R+-DNA and pPB455 is graphed as a function of the total amount of DNA present in the reaction.
Restriction enzymes like many sequence specific enzymes, bind first to non-specific DNA and then transfer to the recognition sequence (2,7,29). Therefore, the peak corresponding to the enzyme–DNA complex in Figure 2A could contain a mixture of enzyme molecules bound to both the recognition sequence as well as to non-specific DNA. To determine the amount of protein bound to non-specific DNA, binding experiments were done using R0-DNA in the absence of a nucleoside cofactor, and the enzyme–DNA mix subjected to gel filtration. The profile demonstrated two well separated peaks as expected, corresponding to the EcoR124I–DNA complex and free protein (data not shown). Analysis of SDS–PAGE and agarose gels of fractions from each peak shows that 71-93% of the protein present is found associated with R0-DNA (Table 1). Identical results were obtained using 2R+-DNA in the absence of nucleoside (Table 1).
Table 1.
Nucleoside cofactors reduce non-specific DNA bindinga
| Reaction |
Amount of protein bound to |
|||
|---|---|---|---|---|
|
2R+-DNA |
R0-DNA |
|||
| Mg2+(%) | Ca2+(%) | Mg2+(%) | Ca2+(%) | |
| No cofactor | 97 | 97 | 71 | 93 |
| ADP | 71 | 70 | 3 | ND |
| ATP | ND | 73 | 11 | 13 |
| ATPγS | ND | 57 | <1 | ND |
aBinding reactions were done as described in Materials and Methods section, in the presence of either 5 mM Mg2+ or Ca2+ ions and 1 mM nucleoside (when present). Thereafter, samples were subjected to native gel filtration in buffer B with Ca(Cl)2 replacing Mg(OAc)2 for binding reactions done in the presence of Ca2+ ions. Typical elution profiles contained two peaks such as those shown in Figure 3A. Following gel filtration, peak fractions were subjected to electrophoresis in agarose and SDS–PAGE gels. The gels were then stained, photographed and analyzed to determine the amount of DNA or enzyme present in each fraction. The amount of protein bound is expressed as a percentage of the total amount of protein detected in the complex and the free enzyme peaks. For each condition, the data are from a single chromatography run. ND, not done.
Previous work showed that ATP influenced the binding specificity of EcoK, a type IA enzyme (23). To determine if nucleoside cofactors affect EcoR124I similarly, we examined the influence of ATP, ADP and ATPγS [a non-hydrolyzable analogue of ATP for this enzyme, (data not shown)] on the interaction of the enzyme with (-)scDNA. The resulting gel filtration elution traces contained two peaks corresponding to the complex and free protein, respectively (Figure 2A and data not shown). Analysis of SDS–PAGE and agarose gels shows that in the presence of ADP or ATPγS, binding to R0-DNA was reduced to ≤3% (Table 1). In contrast, 57–70% of the protein was bound to 2R+-DNA in the presence of these cofactors. In the presence of ATP, binding to R0-DNA was reduced 7- to 9-fold to 12%, relative to nucleoside-free reactions, independent of the metal ion present. As the combination of Mg2+ ions and ATP results in cleavage of site-specific DNA, binding in the absence of cleavage cannot be observed under these conditions. Control assays demonstrate that Ca2+ ions do not support DNA cleavage but do support binding to DNA (data not shown). Consequently, binding assays done in the presence of ATP and Ca2+ ions results in 73% of the protein being bound to 2R+-DNA, 6-fold more than to R0-DNA (Table 1).
Collectively, these data are consistent with a model suggesting that the affinity of EcoR124I for DNA, like that of EcoK, is affected by the presence of a nucleoside cofactor (23). In the absence of nucleoside, affinity for non-specific DNA is high, whereas in the presence of nucleoside cofactors, affinity is altered so that preferential binding to target sequences is facilitated.
EcoR124I is a catalytic enzyme
Using conditions established above, protein–DNA complexes formed on 2R+-DNA in the presence of ADP and Mg2+ were purified using gel filtration. Each fraction of the first peak that eluted from the gel filtration column, that is the protein–DNA complex, was subjected to agarose and SDS–PAGE gel electrophoresis and analysis to determine the ratio of enzyme to DNA present in the complex. The analysis of these gels showed that in the apex fractions, the holoenzyme to site ratio is 1 (Figure 2B).
Then, using the purified stoichiometric EcoR124I–DNA complex, we asked is the enzyme catalytic relative to DNA? To do this, the apex fractions from the complex peak were added to a reaction mix containing ATP and an ATP-regenerating system. Once the reaction had proceeded to completion, a second, distinct-sized R+-DNA (4.3 kB) was added and the reaction continued. Thereafter, time points were subjected to electrophoresis in agarose gels. The results show that the 2R+-DNA (2.9 kB) which was present in the enzyme-DNA complex, was cleaved in <1 min (Phase I, Figure 2C). Significantly, the second (-)scDNA substrate was completely cleaved albeit at a 2-fold reduced rate relative to that of the first DNA (Figure 2C and D). As the only enzyme present in this reaction was that in the purified complex, the only way in which the second DNA could be restricted is if EcoR124I cleaved the first DNA and then transferred from the nascent linear product DNA to the second (-)scDNA resulting in its cleavage. Therefore, EcoR124I is a catalytic enzyme.
Support for enzyme transfer from the linear product is observed when the level of product DNA is quantitated. Analysis of the gel in Figure 2C demonstrates that the nascent linear DNA decreases at a rate of 2.6%/min during phase I (open squares; Figure 2D). Once the second DNA is added, the rate of degradation of the first linear product is reduced 4.3-fold during phase II to 0.6%/min (open circles; Figure 2D). This reduction in cleavage results from EcoR124I transferring from the nascent linear 2R+-DNA to the second (-)scDNA leading to its cleavage. Transfer from product to subsequent substrate is expected for a catalytic enzyme. We note that transfer is not 100% since the linear 2R+-DNA product continues to be degraded. This suggests that the 2R+ substrate is still the favored cleavage substrate with the linear being further degraded to products of nonspecific size.
EcoR124I can perform multiple rounds of DNA cleavage
Thus far, we have shown that EcoR124I can cleave two distinct-sized substrates when added sequentially to an ongoing reaction. As this was demonstrated using a purified enzyme–DNA complex that contained 1 holoenzyme per site, cleavage of the second DNA means that EcoR124I is by definition catalytic. It is conceivable however, that the determination of the holoenzyme to DNA stoichiometry in the complex may be imprecise and there is a sufficient amount of nonspecifically bound protein that transfers from the DNA in the complex to the second DNA leading to its cleavage. Even though we have taken great care to accurately determine the ratio of holoenzyme to sites in the purified complex, there may still be a small of amount of nonspecifically bound enzyme present that has escaped our detection.
If EcoR124I is truly catalytic however, then it should be able to cleave increasingly larger amounts of a second DNA when added to a reaction containing the purified EcoR124I–DNA complex. If the apparent catalytic behavior is due to enzyme bound nonspecifically to the DNA, then the amount of DNA cleaved will saturate at a level corresponding to the amount of total enzyme present in the assay.
To determine which of these possibilities is correct, complexes containing ADP and 2R+-DNA (2.9 kB) were purified and analyzed as before to ascertain the ratio of enzyme to sites (a ratio of 1 was obtained; data not shown). Then, aliquots from the peak fractions were added to separate reaction mixes containing increasing amounts of pPB455 (R+-DNA; 9.8 kB), ATP and an ATP-regenerating system. The final concentration of enzyme in each assay was 6.6 nM. The total DNA concentration in each assay ranged from 10 to 70 nM molecules and consisted of the DNA present in the purified complex (3.3 nM) and additional pPB455.
Following DNA cleavage, samples were subjected to electrophoresis in agarose gels and the resulting gel analyzed. The results show that as the amount of substrate in the reaction increased, the amount of linear product increased as well, with as much as 56 nM being produced from the reaction containing 70 nM total DNA (Figure 2E). As the concentration of active EcoR124I was 6.6 nM, this means that ∼8.5 catalytic cycles were observed. As the only protein present in each assay was that in the purified complex and the amount of linear product produced greatly exceeds the amount of EcoR124I present, each holoenzyme is able to perform multiple, cleavage events. Therefore, we conclude EcoR124I is a catalytic RE.
EcoR124I holoenzyme is a stable complex
Previous work using enzyme reconstituted from the MTase and R-subunits just before use suggests that EcoR124I is unstable resulting from frequent R-subunit loss which was observed to occur during the course of reactions (25,26). These data may have resulted from the inability to achieve stably reconstituted holoenzyme which has been demonstrated to require an overnight incubation (25). Although we have used purified holoenzyme in this work, it is conceivable that this pentameric, R2M2S1 complex is itself unstable and may undergo frequent disassembly during the course of a reaction in a manner analogous to that observed for the reconstituted complexes. Therefore, it is necessary to ascertain whether the purified EcoR124I holoenzyme is a stable pentameric complex. That is, the R2M2S1 complex.
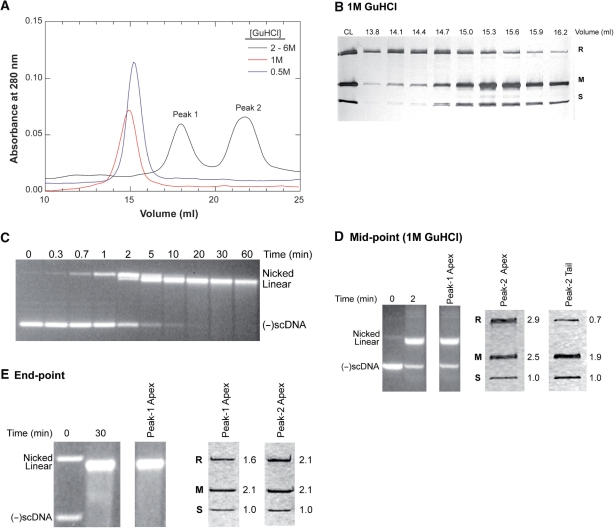
To address this issue, purified holoenzyme was mixed with various concentrations of GuHCl ranging from 0.5 to 6M in separate experiments, and subjected to gel filtration under conditions where the column was equilibrated in running buffer containing the same concentrations of GuHCl. Then, samples were dialyzed and subjected to electrophoresis in SDS-PAGE gels followed by analysis to ascertain the subunit composition of each peak. Results show that concentrations of GuHCl ranging from 2 to 6M dissociate the enzyme into its component subunits producing an elution profile consisting of two peaks. The first contains HsdR, while the second contains HsdM and S (Figure 3A and Supplementary Figure 1A). As the concentration of GuHCl is lowered to 1M and below, the enzyme elutes as a single peak with an apparent MW larger than that of the individual subunits alone, suggesting that it is no longer fully dissociated into its component subunits (Figure 3A). Gel analysis of the peak reveals that in 1 M GuHCl the enzyme is a mixture of free M and R subunits (eluting in the leading edge of the peak), holoenzyme (central fractions) and a complex with an R1M2S1 subunit ratio eluting in the trailing edge of the peak (Figure 3B). Loss of one R-subunit from the holoenzyme is consistent with previous work showing that the two R-subunits bind the MTase with different affinities (25).
Figure 3.
The EcoR124I holoenzyme is stable complex. (A) Denaturing gel filtration elution traces of EcoR124I in the presence of different concentrations of GuHCl. As identical profiles were obtained at [GuHCl] ranging from 2 to 6 M, only a representative trace done at 3 M GuHCl is shown for clarity. (B) A 12% SDS–PAGE gel of dialyzed chromatography fractions from the profile in the presence of 1 M GuHCl shown in (A). The gel was silver stained and photographed immediately; CL, column load. (C) An agarose gel showing DNA cleavage by EcoR124I at 0°C. Reactions contained 5 nM 2R+-DNA and 20 nM active EcoR124I and were initiated by the addition of ATP. Following electrophoresis, gels were stained with ethidium bromide and photographed. (D) Gel analysis of gel filtration peaks from the reaction mid-point from a DNA cleavage assay done at 0°C. Reactions were stopped by the addition of GuHCl (1 M, final) and subjected to native gel filtration. Left, agarose and right, SDS–PAGE gel lanes of gel filtration fractions. The first two agarose lanes contain aliquots from the time course prior to loading onto the column and are presented to demonstrate reaction progress. Due to the presence of 1 M GuHCl, the enzyme dissociates from the DNA and is detected only in the second peak of the elution profile. The apex fraction eluted at 15–15.3 ml and the tail fraction eluted at 16.2 ml (compared to panel B). The values to the right of the SDS–PAGE gel lanes indicate the ratio of each subunit as determined by densitometry with the value for the S-subunit being set to 1 as described previously (11). (E) Gel analysis of the reaction end-point. The reaction mix was loaded directly onto the column. Left, agarose and right, SDS–PAGE gel lanes from fractions from native gel filtration. The first two agarose lanes contain aliquots from the time course prior to loading onto the column and are presented to demonstrate reaction progress. As an excess of enzyme was used, it is detected in both the complex and free protein peaks. The values to the right of the SDS–PAGE gel lanes indicate the ratio of each subunit as determined by densitometry with the value for the S-subunit being set to 1 (11).
In 0.5 M GuHCl, the protein also elutes from the column as a single peak. However, and in contrast to what was observed in 1 M GuHCl, the enzyme is intact as densitometry of each gel lane results in an average R:M:S subunit ratio of 1.7:2.9:1 (Supplementary Figure 1B). Therefore, as concentrations of GuHCl ≥ 1 M are required to completely dissociate the holoenzyme into its component subunits, we conclude that in the absence of DNA and ATP, the purified EcoR124I holoenzyme is a stable complex.
Holoenzyme stability is observed during the course of a reaction
Although the EcoR124I holoenzyme is an intrinsically stable complex, it is conceivable that during the course of a reaction it can still dissociate into its component subunits and reassociate in solution to form the holoenzyme. Therefore, we chose two time course checkpoints at which to measure complex stability and challenged the enzyme with GuHCl. The mid-point was selected as the enzyme may disassemble during the course of a reaction. As dissociation may occur once the cleavage reaction has completed and nascent DNA ends are released, we evaluated enzyme stability at the reaction end-point as well.
DNA cleavage assays were performed on ice (i.e. 0°C) so that the mid-point could be precisely obtained. At this temperature, 50% of the (-)scDNA substrate is cleaved in 2 min with the reaction completed by 30 min (Figure 3C). Reactions were terminated at the mid-point by the addition of GuHCl to 1 M final, and the entire mix subjected to native gel filtration. As this concentration of GuHCl partially dissociates the holoenzyme, and if EcoR124I dissociates during the course of a reaction, the presence of 1 M GuHCl may further destabilize the enzyme, producing an elevated level of dissociated R-subunit relative to that shown in Figure 3B. The resulting elution profile showed two peaks as expected; the first contains DNA only while the second contains protein only (data not shown). The agarose gel of the first peak shows that 50% of the input DNA was cleaved as expected (Figure 3D; left panel). Analysis of the protein gel of the second peak revealed no additional dissociation of the R-subunit beyond that observed in the absence of DNA (Figure 3D; right panel; compare to Figure 3B).
Identical experiments were done to ascertain enzyme stability following reaction completion (i.e. at 30 min; Figure 3C). Thereafter, reaction mixes were loaded directly onto the column. As for the mid-point, no detectable dissociation of the holoenzyme either still bound to the DNA (peak 1) or free in solution (peak 2), was observed (Figure 3E, right panel). An additional experiment was done where GuHCl was added to 1 M final at 30 min, and the mixture loaded onto the column. Here, limited R-subunit dissociation was observed as shown in Figure 3B and D, and as for the mid-point, there was no enhancement in R-subunit dissociation relative to that observed in the absence of DNA and ATP (data not shown). Thus, within the limits of detection in this assay, the holoenzyme does not appreciably dissociate into its component parts during the course of a reaction. Therefore, we conclude that when purified as the holoenzyme, EcoR124I is a stable, pentameric complex.
EcoR124I is active as a dimer of holoenzymes
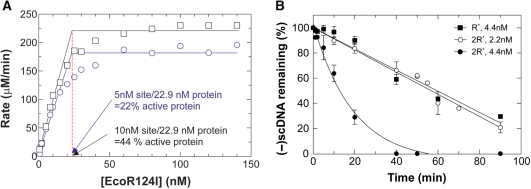
To determine the fraction of active enzyme in our protein preparations, we utilize a coupled, spectrophotometric ATPase assay, titrate total protein relative to negatively supercoiled, R+-DNA and measure the initial rates of ATP hydrolysis (11). The data show that as total protein concentration increases, the initial rates of ATP hydrolysis increase accordingly (Figure 4A). As EcoR124I does not appreciably hydrolyze ATP in the absence of DNA or in the presence of R0-DNA (11), the increased levels of activity correlate with occupancy of target sequences by active holoenzyme present in the total protein population. For this preparation, the data show that activity saturated at a concentration of 22.9 nM protein, indicating that 22% of the protein present was active. This is only marginally higher than 16% active protein reported previously (11).
Figure 4.
EcoR124I is active as a dimer of holoenzymes. (A) The active fraction of enzyme is influenced by the number of recognition sites present in the DNA substrate. A titration of protein relative to (-)scDNA was done as described in Materials and Methods section and the rates of ATP hydrolysis determined. To accurately determine the intercept for stoichiometry, the lines fit to the data prior to saturation were determined by linear regression (a minimum of seven data points were used in the fitting to each data set). Each data set is from two separate assays with each assay containing the alternate concentrations of protein. Circles, R+-DNA; squares, 2R+-DNA. (B) EcoR124I is active as a dimer. The analysis is of agarose gels from DNA cleavage assays done in the presence of either R+-(squares) or 2R+-DNA (circles). Reactions were done at 0°C and contained 5 nM DNA and EcoR124I at the concentrations shown. The amount of (-)scDNA remaining at each time point is expressed as a fraction of the input. The data represent three independent time courses done on separate days.
The low level of active enzyme is troubling since preparations are >95% homogenous and contain stoichiometric ratios of the R-, M- and S-subunits (2:2:1; data not shown and (11)). The reason for this low level of active enzyme is unknown. To determine whether additional factors might be limiting, we repeated the protein titrations relative to R+-DNA in the presence of different concentrations of Mg[OAc]2 or SAM, in separate experiments. Similarly, we also varied the concentration of Mg[OAc]2 or SAM while holding the remaining components constant. The results show that within experimental error, the fraction of active enzyme remained unaffected (data not shown). Next, the protein titration was repeated using negatively supercoiled 2R+-DNA instead of R+-DNA. We expected that activity would saturate at 46 nM total protein. Surprisingly, saturation was observed at 22.9 nM protein indicating that the level of active protein had doubled to 44% (Figure 5A). These data suggest that EcoR124I functions as a dimer of holoenzymes consistent with previous predictions for other Type I enzymes (5,30).
Figure 5.
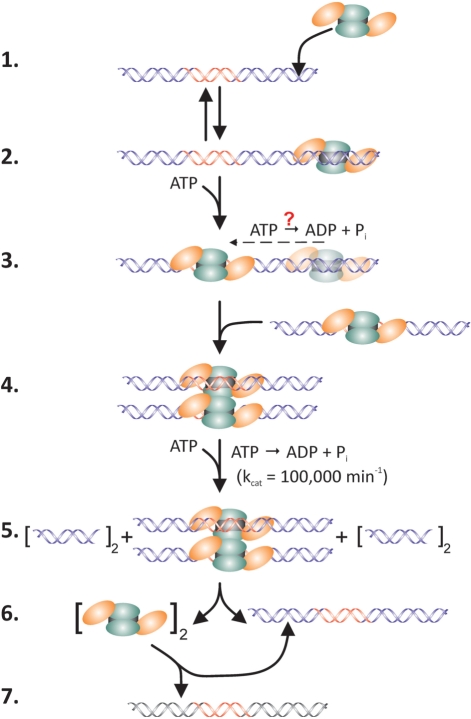
The catalytic reaction mechanism of EcoR124I. (1) The holoenzyme binds to non-specific sites on the DNA forming the initial complex. (2) ATP binds to the enzyme facilitating transfer from non-specific DNA to the target sequence (indicated in red). Hydrolysis may occur and involve either one or both R-subunits to produce the recognition complex. (3) A second recognition complex binds to the first, forming the active dimer. (4) Rapid ATP hydrolysis coupled to dsDNA translocation ensues. Loops form due to translocation of DNA towards the stationary holoenzymes bound to their target sequences (data not shown). (5) Translocation is impeded leading to sequential DNA nicking producing dsDNA breaks. (6) Holoenzymes dissociate from the restricted DNA. The subscript 2 indicates two holoenzymes; dimers are not implied. (7) Intact holoenzymes bind to either the nascent product or to a subsequent substrate DNA if present (black/red DNA). Orange ovals, R-subunits; green ovals, M-subunits and black rectangle, S-subunit.
Previous work demonstrated a sigmoid, non-linear dependence of the rate of DNA cleavage on the concentration of total EcoR124I protein (9). (In this work, the fraction of active enzyme was not determined.) These findings are consistent with the requirement for dimer formation with the possibility of cooperative protein-protein interactions being involved. To further investigate this nonlinear dependence, we varied the active enzyme concentration from 1 to 8 nM in the presence of 5 nM R+-DNA and measured the reaction rate. A similar, nonlinear dependence of DNA cleavage rate on enzyme concentration was observed (data not shown).
To examine the nonlinear dependence more clearly, we conducted a careful comparison of two concentrations of active enzyme from the abovementioned titration. Concentrations of 2.2 and 4.4 nM active enzyme were chosen, corresponding to ∼50% saturation and near saturation of the 5 nM R+-DNA substrate (Figure 4A). DNA cleavage assays were done as described in Materials and methods section and the analysis of the resulting gels is shown in Figure 4B.
At 2.2 nM active enzyme, 5 nM DNA was cleaved at a rate of 0.9%/min with 80% of the input DNA cleaved in the 90 min assay. The kcat for this reaction was determined to be 0.035 min−1. When the concentration of active enzyme was increased to 4.4 nM, the DNA cleavage rate increased 3.9-fold to 3.6%/min and 100% of the substrate was cleaved by 40 min. The resulting kcat was 0.142 min−1, 4-fold higher than the 2.2 nM enzyme reaction.
Next, we repeated the DNA cleavage assay using the higher concentration of active enzyme (i.e. 4.4 nM), reduced the number of sites present in the (-)scDNA substrate from two to one while holding the concentration of DNA molecules constant at 5 nM. We reasoned that if the enzyme is a dimer, then by placing sites in separate DNA molecules the probability of forming the dimer should be reduced and this might be observed as a decreased rate of cleavage. As anticipated, the results show that R+-DNA was cleaved at a rate of 0.84%/min, down 4.3-fold compared to the reaction containing 2R+-DNA (Figure 4B).
The nonlinear dependence of the reaction rate on active enzyme concentration and the quadrupling of the reaction rate that occurs when either the active enzyme concentration or the number of sites present in the substrate is doubled, are consistent with the requirement for dimer formation. As EcoR124I has no demonstrable activity in the absence of DNA or in the presence of R0-DNA (11), these data suggest that the active form of the protein is a dimer with each holoenzyme simultaneously bound to DNA containing a recognition sequence. Clearly, additional work is required to further elucidate dimerization. However, these solution results are consistent with previous work for both EcoR124I and EcoK and with previous predictions for the dimerization requirement for type I REs (5,9,30).
DISCUSSION
The primary conclusion of this work is that type I REs are true enzymes. That is, they act catalytically with respect to DNA, ATP and SAM. This is the first demonstration of complete catalytic behavior for any type I RE. The data also show that the active form of EcoR124I is a dimer of holoenzymes, with each bound to a recognition sequence. These results go hand-in-hand to explain how these enzymes function.
For the past 35 years, type I REs have existed as an enigma (1,5). This view has held as the enzymes bind to sequence-containing DNA, rapidly hydrolyze ATP, cleave the dsDNA in cis and then were thought to remain bound to that same DNA. In this state they remain as oxymoronic, stoichiometric ‘enzymes’ incapable of acting on additional DNA molecules.
In this work, we show for the first time that type I REs are instead, true enzymes, functioning catalytically with respect to DNA. The unequivocal demonstration of this point required removal of free protein and purification of the holoenzyme bound to ADP and a DNA substrate containing the recognition sequence. Purified stoichiometric complexes (i.e. 1 holoenzyme per DNA site) were added to a reaction mix and once cleavage was complete, a second, distinct-sized R+-DNA was added. The data show that both DNA substrates were cleaved. As the only enzyme present was that bound to the DNA in the complex, and both DNA molecules in the reaction were cleaved, we conclude that EcoR124I is catalytic relative to DNA. If the enzyme were stoichiometric, the second DNA substrate could never be cleaved. To confirm catalytic function, we added gel filtration-purified, stoichiometric enzyme–DNA complexes to reactions containing increasingly greater amounts of a second DNA (up to a combined total of 70 nM DNA molecules). The data showed that EcoR124I was able to cleave as much as 80% of the total DNA, undergoing approximately eight catalytic cycles. Therefore, we conclude that EcoR124I is indeed catalytic. Consequently, type I REs are true enzymes.
The catalytic function observed here is not due to frequent disassembly of the EcoR124I holoenzyme into methyltransferase and R-subunits as proposed previously from studies using reconstituted enzyme (25,26). This was demonstrated by challenging the purified holoenzyme with increasing concentrations of the denaturant, guanidinium hydrochloride. The results show that the purified holoenzyme remained intact in up to 0.5 M GuHCl and furthermore, complex stability persisted during the course of reactions. Therefore, any model that attributes subunit association and dissociation as a means to control enzyme activity cannot apply to the EcoR124I holoenzyme (26). Furthermore, these results indicate that once EcoR124I has translocated and cleaved one DNA molecule, it transfers as an intact, R2M2S1 entity to subsequent DNA substrates, leading to their cleavage.
Protein titrations in ATPase assays were used to determine the level of active protein. In addition to providing the active stoichiometry, these data demonstrated that when the number of binding sites in the (-)scDNA substrate was doubled, the fraction of active protein unexpectedly doubled as well, suggesting the requirement for a dimer (Figure 4A). Further investigation of the dependence of DNA cleavage reaction rate on concentration of active enzyme proved insightful. Initial experiments revealed a sigmoid, nonlinear dependence of reaction rate on enzyme concentration, consistent with previous work from other laboratories (9). Careful analysis in this work revealed that when the concentration of active enzyme was doubled in the presence of 2R+-DNA, the reaction rate quadrupled (Figure 4B). If the enzyme were active as a monomer (i.e. a single R2M2S1 complex), then doubling the concentration of enzyme should result in the reaction rate simply doubling. This is clearly not what was observed. Instead the results are more complex and are consistent with the active form of the enzyme being a dimer of holoenzymes, i.e. two R2M2S1 complexes with each bound to a recognition site simultaneously. Each holoenzyme must be bound to a target sequence simultaneously since no activity is observed in the absence of DNA or in the presence of R0-DNA (11). The data are consistent with a previous model suggesting dimerization is intrinsic to type I RE function (5). Dimerization is not limited to EcoR124I having been observed for other type I holoenzymes (17,18,30). For EcoKI, dimerization prior to the onset of dsDNA translocation was observed using atomic force microscopy. Although these images were done in the absence of ATP, dimerization required occupancy of recognition sequences by each multi-subunit member of the dimer (30). Furthermore, dimerization is also a requirement for the unusual type II RE, Fok I (31).
Intriguingly, the requirement for a dimer also partially explains the apparent saturation behaviour observed in enzyme titrations (Figure 4A). For a stoichiometric enzyme, increasing the amount of total protein relative to a fixed amount of substrate yields a linear increase in activity which saturates when all of the available substrate is bound. Once saturation is achieved, further increases in protein do not produce further increases in activity. This was observed for EcoR124I in both DNA cleavage (data not shown) and ATPase assays (Figure 4A). It is now clear that these titrations do not reflect true stoichiometric behaviour but instead reflect the dimerization requirement and is related to occupancy of recognition sites. At low concentrations of protein, the probability of forming a dimer is rare so that very little activity is observed. As protein concentration increases, the probability of dimer formation increases and correspondingly, initial rates of activity do as well. Once all sites are occupied, the probability of dimer formation is maximal and consequently, reactions rates are as well. Intriguingly, we consistently observe elevated higher rates of activity in the presence of 2R+- relative to R+-DNA at the same concentrations of active enzyme. In fact, the rate of DNA cleavage is typically 4-fold higher (Figure 4B). This result is also consistent with the requirement for dimerization as intra-molecular dimers (i.e. bound to two sites present on the same DNA) are more likely to form than inter-molecular ones (i.e. holoenzymes bound to separate DNA molecules as they would be with single site-containing DNA).
The requirement for a dimer is not the full story however. Full catalytic activity (i.e. 100% cleavage of all available substrate) was only observed for EcoR124I when an enzyme to DNA ratio of approximately 1 was used (Figure 1 and data not shown). This is surprising as an enzyme should be catalytic regardless of protein concentration. The failure to observe catalytic function at low concentrations of protein is readily explained when one considers that the recognition sequences present in the substrate are neither destroyed nor inactivated during the course of a cleavage reaction. Recall that cleavage by type I enzymes occurs at sites distant from the recognition sequence and not within or immediately adjacent to the sequence as in the case of type II enzymes (7,16). Therefore, during the course of a reaction, there is a constant redistribution of enzyme by the available sites present in both the substrate and product DNA molecules. As full cleavage activity requires formation of a dimer bound to two substrate–DNA target sequences simultaneously, the nascent linear DNA product acts as a competitor molecule making it increasingly more difficult to form the active dimer (i.e. bound to two substrate DNAs). This competition which increases as the reaction progresses, results in the appearance of stoichiometric behaviour. The inability to completely cleave all available substrate is further complicated by the presence of free protein, some of which is inactive (the level of inactive protein is currently unknown). When free protein was removed by gel filtration, the ‘purified’ fraction of enzyme was consistently more active as indicated by higher rates and extents of reaction compared to the initial partially active pool of enzyme (data not shown).
The product inhibition model predicts that if the linear product were removed from the reaction, EcoR124I should demonstrate full catalytic function, even at low concentrations of active enzyme relative to DNA. This was shown previously in vitro when cleavage assays were done in the presence of the potent DNA helicase, the E. coli RecBCD enzyme, which exhibits a >106-fold higher affinity for linear dsDNA relative to closed circular forms (32). Here, sub-stoichiometric amounts of EcoR124I were able to cleave subsequent (-)scDNA substrate molecules added to reactions only when RecBCD was present but not in its absence (11). The original purpose of the combined EcoR124I-RecBCD assay was to determine whether EcoR124I released the nascent DNA ends immediately following DNA cleavage. As RecBCD was able to rapidly unwind EcoR124I-restricted dsDNA, we concluded that ends were being released. We then added a second aliquot of (-)scDNA to this reaction following the unwinding by RecBCD and found that to our surprise, it too was restricted and rapidly unwound. As we were unaware that EcoR124I acted in catalytic fashion, we proposed that (i) the initial DNA cleavage event somehow ‘inactivated’ EcoR124I and (ii), displacement by RecBCD then ‘reactivated’ EcoR124I resulting in cleavage of subsequent DNA molecules (11). We now know that this was incorrect and the observed effect was instead due to removal of the competitor, linear product DNA by RecBCD thereby enabling EcoR124I to facilitate multiple rounds of DNA cleavage.
These in vitro results are consistent with previous in vivo data demonstrating a reduction in restriction by EcoKI in the absence of RecBCD (33). Although the reason for this was unclear at the time, our data suggest that the reduction may at least be due in part to the accumulation of competitor DNA resulting from DNA cleavage by EcoKI. As RecBCD was unavailable to degrade restricted DNA, EcoKI could re-bind these DNA molecules and consequently was unable to cleavage additional DNA. In the presence of RecBCD, the linear DNA is effectively removed allowing EcoKI to cleave additional DNA. Further support for the coupling between nucleases and REs comes from work with R.EcoP151, a type III RE (34). In the absence of an exonuclease, EcoP15I was only able to perform a single round of DNA cleavage. In contrast, in the presence of Exonuclease III, R.EcoP151 was able to perform multiple rounds of DNA cleavage. Taken together, these findings suggest that for enzymes such as type I and III REs that do not destroy their target sequence during restriction, removal of the nascent competitor product DNA is a previously underappreciated and key component of the reaction pathway. Failure to do so limits the restriction enzyme to a single round of cleavage whereas the presence of the exonuclease ensures catalytic function by removal of inhibitor product DNA.
As EcoR124I is a stable multi-subunit enzyme, the following catalytic reaction scheme emerges (Figure 5). The stages up to cleavage of the dsDNA substrate (excluding conformational changes induced by SAM) are similar to those proposed previously for EcoK, thus the scheme presented should apply to all type I enzymes (8). The intact, monomeric holoenzyme binds first to nonspecific DNA, forming the initial complex (steps 1 and 2). In the presence of a nucleoside cofactor, the enzyme transfers to a nonmethylated recognition sequence embedded within dsDNA, forming the recognition complex (step 3). Transfer may involve hydrolysis of a few molecules of ATP, since nonspecific binding is reduced in the presence of ADP, although more work is required to elucidate the nucleoside requirement more precisely (Table 1). Once at the recognition sequence, dimerization occurs (step 4). This is presented schematically in only one way for clarity with two, monomeric EcoR124I–DNA complexes interacting. Alternatively, formation of the dimer may occur when a second EcoR124I holoenzyme collides with the first already bound to a recognition sequence. This would then be followed by binding of a second DNA. Once the complete dimer has formed and, if the DNA is unmethylated, it is followed by rapid, ATP hydrolysis-driven dsDNA translocation leading to restriction of the DNA at a position distant from the recognition sequence (step 5). Immediately thereafter, the intact holoenzymes transfer from the nascent cleaved DNA to subsequent DNA molecules (if present), and the process is repeated (steps 6 and 7). If a second DNA molecule is absent, the enzymes rebind to the nascent linear DNA product (which still contains the intact recognition sequence) and proceed to hydrolyze ATP and nonspecifically degrade the product to which it has rebound (step 6).
Rebinding to the linear product is reduced once subsequent substrate DNA molecules are added to the reaction (Figure 2D and E). Further support for rebinding of the product DNA comes from several studies where post-cleavage, futile cycles of ATP hydrolysis have been observed to persist for several hours (11,22,35,36). In addition, a previous characterization of the ATPase activity of EcoR124I revealed that in the presence of (-)scDNA, time courses are biphasic with the first rapid phase corresponding to hydrolysis prior to DNA cleavage and the second 10-fold slower phase corresponding to post-cleavage hydrolysis (11). When heparin was added along with ATP following binding of the enzyme to DNA, the first phase of ATP hydrolysis was unaffected while the second phase was eliminated. This occurs because the enzyme dissociates from the nascent linear product and binding to additional DNA substrates is inhibited as the enzyme is sequestered by the heparin. When heparin is added prior to protein, all type I activities are inhibited (8,11).
The observation of catalytic function for type I REs alters the in vivo mechanistic view of these previously enigmatic enzymes. The idea that these enzymes might remain bound to restricted phage DNA suggests a requirement for their rapid removal by either the action of exonucleases such as RecBCD or ExoIII (11,33,34) or by proteases (37–39). Failure to remove type I restriction enzymes from the DNA using the abovementioned, ATP-requiring processes either separately, or in combination, could prove costly to a cell. This follows because the prolific ATPase activity (kcat = 100 000 min−1) of these enzymes could potentially deplete the intracellular ATP pools leading to cell death as suggested previously (24). As type I REs release from the DNA following restriction, these enzymes do not pose a threat to the survival of the infected host following restriction of invading DNA. Instead they utilize a unique reaction mechanism to recognize and respond to the methylation state of target sequences embedded within dsDNA and take advantage of the supply of ATP to rapidly restrict invading DNA to the benefit of the host. The restricted DNA is a substrate for exonucleases such as RecBCD and the combined actions of the R–M and recombination machinery results in effective annihilation of invading DNA (11,40). This leaves the catalytic EcoR124I holoenzyme free to act on subsequent invading DNA molecules in the absence of competitor, product DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM66831). Funding for open access charge: National Institutes of Health GMS Grant GM66831.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Hui and Meixia Gao for reading the article.
Footnotes
Present address: Cuiling Xu, Department of Biochemistry, 726 Biological Science Building, 484 West 12th Ave., The Ohio State University, Columbus, OH 43212, USA
REFERENCES
- 1.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sistla S, Rao DN. S-Adenosyl-L-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:1–19. doi: 10.1080/10409230490440532. [DOI] [PubMed] [Google Scholar]
- 3.Bickle TA. Restricting restriction. Mol. Microbiol. 2004;51:3–5. doi: 10.1046/j.1365-2958.2003.03846.x. [DOI] [PubMed] [Google Scholar]
- 4.Dryden DT, Murray NE, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourniquel AA, Bickle TA. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84:1047–1059. doi: 10.1016/s0300-9084(02)00020-2. [DOI] [PubMed] [Google Scholar]
- 6.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Modrich P. Studies on sequence recognition by type II restriction and modification enzymes. CRC Crit. Rev. Biochem. 1982;13:287–323. doi: 10.3109/10409238209114231. [DOI] [PubMed] [Google Scholar]
- 8.Yuan R, Bickle TA, Ebbers W, Brack C. Multiple steps in DNA recognition by restriction endonuclease from E. coli K. Nature. 1975;256:556–560. doi: 10.1038/256556a0. [DOI] [PubMed] [Google Scholar]
- 9.Janscak P, Abadjieva A, Firman K. The type I restriction endonuclease R.EcoR124I: over-production and biochemical properties. J. Mol. Biol. 1996;257:977–991. doi: 10.1006/jmbi.1996.0217. [DOI] [PubMed] [Google Scholar]
- 10.Davies GP, Martin I, Sturrock SS, Cronshaw A, Murray NE, Dryden DT. On the structure and operation of type I DNA restriction enzymes. J. Mol. Biol. 1999;290:565–579. doi: 10.1006/jmbi.1999.2908. [DOI] [PubMed] [Google Scholar]
- 11.Bianco PR, Hurley EM. The type I restriction endonuclease EcoR124I, couples ATP hydrolysis to bidirectional DNA translocation. J. Mol. Biol. 2005;352:837–859. doi: 10.1016/j.jmb.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 12.Mernagh DR, Janscak P, Firman K, Kneale GG. Protein-protein and protein-DNA interactions in the type I restriction endonuclease R.EcoR124I. Biol. Chem. 1998;379:497–503. doi: 10.1515/bchm.1998.379.4-5.497. [DOI] [PubMed] [Google Scholar]
- 13.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Ann. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 14.Studier FW, Bandyopadhyay PK. Model for how type I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horiuchi K, Zinder ND. Cleavage of bacteriophage fl DNA by the restriction enzyme of Escherichia coli B. Proc. Natl Acad. Sci. USA. 1972;69:3220–3224. doi: 10.1073/pnas.69.11.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreier J, MacWilliams MP, Bickle TA. DNA cleavage by the type IC restriction-modification enzyme EcoR124II. J. Mol. Biol. 1996;264:722–733. doi: 10.1006/jmbi.1996.0672. [DOI] [PubMed] [Google Scholar]
- 18.Janscak P, MacWilliams MP, Sandmeier U, Nagaraja V, Bickle TA. DNA translocation blockage, a general mechanism of cleavage site selection by type I restriction enzymes. EMBO J. 1999;18:2638–2647. doi: 10.1093/emboj/18.9.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- 20.Jindrova E, Schmid-Nuoffer S, Hamburger F, Janscak P, Bickle TA. On the DNA cleavage mechanism of Type I restriction enzymes. Nucleic Acids Res. 2005;33:1760–1766. doi: 10.1093/nar/gki322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan R, Heywood J, Meselson M. ATP hydrolysis by restriction endonuclease from E. coli K. Nature. 1972;240:42–43. doi: 10.1038/newbio240042a0. [DOI] [PubMed] [Google Scholar]
- 22.Eskin B, Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. II. Purification, subunit structure, and catalytic properties of the restriction endonuclease. J. Biol. Chem. 1972;247:6183–6191. [PubMed] [Google Scholar]
- 23.Bickle TA, Brack C, Yuan R. ATP-induced conformational changes in the restriction endonuclease from Escherichia coli K-12. Proc. Natl Acad. Sci. USA. 1978;75:3099–3103. doi: 10.1073/pnas.75.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bickle TA. In: Nucleases. 2nd. Linn SM, Lloyd RS, Roberts RJ, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 89–109. [Google Scholar]
- 25.Janscak P, Dryden DT, Firman K. Analysis of the subunit assembly of the typeIC restriction-modification enzyme EcoR124I. Nucleic Acids Res. 1998;26:4439–4445. doi: 10.1093/nar/26.19.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidel R, Bloom JG, van Noort J, Dutta CF, Dekker NH, Firman K, Szczelkun MD, Dekker C. Dynamics of initiation, termination and reinitiation of DNA translocation by the motor protein EcoR124I. EMBO J. 2005;24:4188–4197. doi: 10.1038/sj.emboj.7600881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. [erratum appears in Anal Biochem 1990 Sep;189:283] Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Rao DN, Saha S, Krishnamurthy V. ATP-dependent restriction enzymes. Progr. Nucleic Acid Res. Mol. Biol. 2000;64:1–63. doi: 10.1016/s0079-6603(00)64001-1. [DOI] [PubMed] [Google Scholar]
- 30.Berge T, Ellis DJ, Dryden DT, Edwardson JM, Henderson RM. Translocation-independent dimerization of the EcoKI endonuclease visualized by atomic force microscopy. Biophysical J. 2000;79:479–484. doi: 10.1016/S0006-3495(00)76309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roman LJ, Kowalczykowski SC. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry. 1989;28:2863–2873. doi: 10.1021/bi00433a018. [DOI] [PubMed] [Google Scholar]
- 33.Salaj-Smic E, Marsic N, Trgovcevic Z, Lloyd RG. Modulation of EcoKI restriction in vivo: role of the lambda Gam protein and plasmid metabolism. J. Bacteriol. 1997;179:1852–1856. doi: 10.1128/jb.179.6.1852-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raghavendra NK, Rao DN. Functional cooperation between exonucleases and endonucleases––basis for the evolution of restriction enzymes. Nucleic Acids Res. 2003;31:1888–1896. doi: 10.1093/nar/gkg275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan R, Hamilton DL, Hadi SM, Bickle TA. Role of ATP in the cleavage mechanism of the EcoP15 restriction endonuclease. J. Mol. Biol. 1980;144:501–519. doi: 10.1016/0022-2836(80)90334-4. [DOI] [PubMed] [Google Scholar]
- 36.Dreier J, Bickle TA. ATPase activity of the type IC restriction-modification system EcoR124II. J. Mol. Biol. 1996;257:960–969. doi: 10.1006/jmbi.1996.0215. [DOI] [PubMed] [Google Scholar]
- 37.Blakely G, Murray NE. Control of the endonuclease activity of type I restriction-modification systems is required to maintain chromosome integrity following homologous recombination. Mol. Microbiol. 2006;60:883–893. doi: 10.1111/j.1365-2958.2006.05144.x. [DOI] [PubMed] [Google Scholar]
- 38.Doronina VA, Murray NE. The proteolytic control of restriction activity in Escherichia coli K-12. Mol. Microbiol. 2001;39:416–428. doi: 10.1046/j.1365-2958.2001.02232.x. [DOI] [PubMed] [Google Scholar]
- 39.Makovets S, Titheradge AJ, Murray NE. ClpX and ClpP are essential for the efficient acquisition of genes specifying type IA and IB restriction systems. Mol. Microbiol. 1998;28:25–35. doi: 10.1046/j.1365-2958.1998.00767.x. [DOI] [PubMed] [Google Scholar]
- 40.Brammar WJ, Murray NE, Winton S. Restriction of lambda trp bacteriophages by Escherichia coli K. J. Mol. Biol. 1974;90:633–647. doi: 10.1016/0022-2836(74)90529-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.