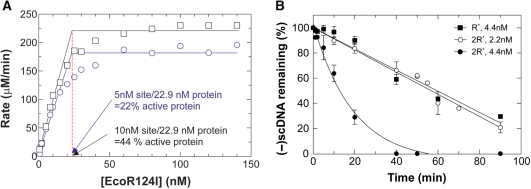
Figure 4.
EcoR124I is active as a dimer of holoenzymes. (A) The active fraction of enzyme is influenced by the number of recognition sites present in the DNA substrate. A titration of protein relative to (-)scDNA was done as described in Materials and Methods section and the rates of ATP hydrolysis determined. To accurately determine the intercept for stoichiometry, the lines fit to the data prior to saturation were determined by linear regression (a minimum of seven data points were used in the fitting to each data set). Each data set is from two separate assays with each assay containing the alternate concentrations of protein. Circles, R+-DNA; squares, 2R+-DNA. (B) EcoR124I is active as a dimer. The analysis is of agarose gels from DNA cleavage assays done in the presence of either R+-(squares) or 2R+-DNA (circles). Reactions were done at 0°C and contained 5 nM DNA and EcoR124I at the concentrations shown. The amount of (-)scDNA remaining at each time point is expressed as a fraction of the input. The data represent three independent time courses done on separate days.