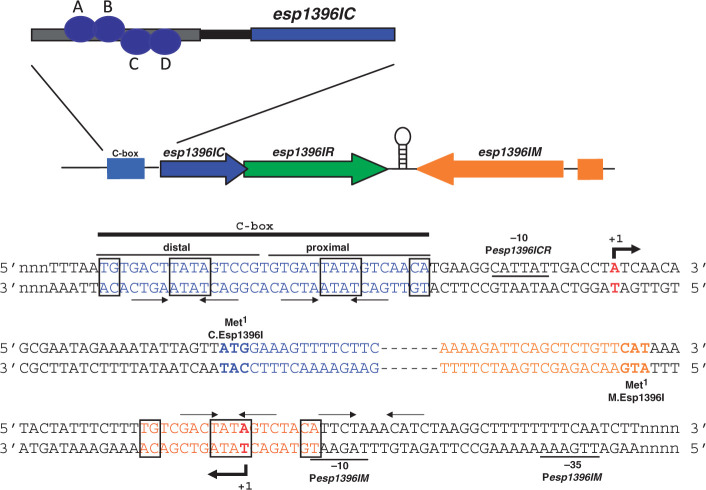
Figure 1.
Genetic organization of restriction–modification system Esp1396I. The esp1396I genes with upstream C.Esp1396I binding sites are schematically shown on the top. Arrows show the direction of transcription. Two esp1396I transcription units (M and CR) are separated by a region that may contain a bidirectional transcription terminator (depicted as a hairpin). DNA sequences around transcription start points of the esp1396IM and esp1396ICR promoters are expanded below (both DNA strands are shown). Beginnings of coding regions are indicated with colors that match those at the top of the figure and esp1396IC and esp1396IM are highlighted in bold typeface and indicated as ‘Met1’. The Pesp1396IM and Pesp1396ICR start sites are indicated by red color and bold typeface, and leftward and rightward arrows below and above the sequence. Conserved promoter elements are labeled and underlined. The esp1396ICR C-box is indicated by blue color and a black line above the sequence with distal and proximal C.Esp1396I binding sites labeled. Two sets of inverted repeats are shown by convergent arrows. Sequences constituting the recently discovered ‘TATA’-based dyad are boxed. Sequences within one set of perfect inverted repeats in Pesp1396IM are similarly indicated and highlighted with orange-colored font. An imperfect set of repeats in Pesp1396IM identified in ref. (8) is shown by inverted arrows above black-colored font. At the top of the figure, the orientation of C.Esp1396I dimers at promoter-distal (subunits A, B) and promoter-proximal (subunits C, D) Pesp1396ICR binding sites is shown (19).