Abstract
Recent metagenomics studies of environmental samples suggested that microbial communities are much more diverse than previously reported, and deep sequencing will significantly increase the estimate of total species diversity. Massively parallel pyrosequencing technology enables ultra-deep sequencing of complex microbial populations rapidly and inexpensively. However, computational methods for analyzing large collections of 16S ribosomal sequences are limited. We proposed a new algorithm, referred to as ESPRIT, which addresses several computational issues with prior methods. We developed two versions of ESPRIT, one for personal computers (PCs) and one for computer clusters (CCs). The PC version is used for small- and medium-scale data sets and can process several tens of thousands of sequences within a few minutes, while the CC version is for large-scale problems and is able to analyze several hundreds of thousands of reads within one day. Large-scale experiments are presented that clearly demonstrate the effectiveness of the newly proposed algorithm. The source code and user guide are freely available at http://www.biotech.ufl.edu/people/sun/esprit.html.
INTRODUCTION
The latest development of massively parallel pyrosequencing technology enables ultra-deep sequencing of complex microbial populations rapidly and inexpensively (1,2). For example, 454 Life Sciences GS FLX systems (Branford, CT, USA) (3) can finish a full pyrosequencing run with more than 400K sequences within one day of operation (http://www.454.com/products-solutions/system-features.asp). It allows researchers to study genetic materials recovered directly from environmental samples, bypassing the needs for isolation and lab cultivation of individual species, and thus opens a new window to probe the hidden world of microbial communities. This technique has been successfully used in several 16S rRNA-based metagenomics analyses of various environments. For example, Sogin et al. (4) provided one of the first global indepth descriptions of microbial diversities and their relative abundance in the ocean, and Keijser et al. (5) were among the first to study oral microbial populations. It has been shown that the microbial diversities are at least one order of magnitude larger than previously reported. These estimation results, however, were computed through extrapolation. In order to obtain more accurate estimates, surveys that are several orders of magnitude larger than those reported in the literature may be required to uncover sequences from minor components (4,5). However, analyzing large collections of 16S ribosomal sequences poses a serious computational challenge for existing algorithms.
In this article, we focus on taxonomy independent analysis where sequences are classified into operational taxonomic units (OTUs) of specified sequence variations, based on which various ecological metrics are estimated. Typically, sequences with <3% dissimilarity are assigned to the same species, while those with <5% dissimilarity are assigned to the same genus, although these distinctions are controversial (6,7,8). One outstanding challenge of taxonomy independent analysis is the alignment of sequences of a sample for the calculation of sequence variations. One commonly used method in the literature is multiple sequence alignment (MSA) [see, for example, (4,5,9)]. While significant improvement has been made in the last decade to reduce the computational complexity of MSA [e.g. MUSCLE (10) and MAFFT (11)], it is still computationally intractable to align hundreds of thousands of sequences. Moreover, the use of MSA for aligning hypervariable regions of 16S rRNA gene has not been well justified in the literature. MSA is used to infer homologous segments of input sequences. An underlying assumption is that input sequences should share some similarities, which may not be valid for 16S rRNA-based studies that target on hypervariable regions of rRNA genes (e.g. V6 and V3 regions). We experimentally found through a benchmark study that the use of MSA leads to an inflated estimate of genetic distances and microbial diversities (see Results Section and Section 1S in the Supplementary Data for a detailed discussion). Another major issue of taxonomy-independent analysis is the high computational complexity and memory requirement associated with assigning sequences into OTUs. DOTUR (8) is a commonly used algorithm for this purpose. It conveniently integrates the tasks of performing clustering and statistical interference of species richness into one program, which frees researchers from manually manipulating a large distance matrix. However, DOTUR does not scale well to handle extremely large data sets. The recently released MOTHUR significantly improved the computational performance of DOTUR. As with DOTUR, MOTHUR loads a distance matrix into memory before proceeding to perform clustering. Hence, it does not fundamentally address the computational issue associated with processing massive pyrosequencing data. Given a full run of 454 data, a full distance matrix can be as large as 1000 GB. Even if we remove duplicated sequences and sequence pairs that have a pairwise distance larger than a cutoff value (say 0.1), the resulting distance matrix in a sparse format can be 20 GB in size, which is too big to be directly loaded into the memory in most computers. There exist a few algorithms that used various strategies to overcome the above mentioned computational issues. Two typical methods are FastGroupII (12) and RDP-Pyro (13). By submitting data to their web applications and through personal communications, we found that these methods can work efficiently only for small- or medium-scale data sets.
In this article, we proposed a new algorithm, referred to as ESPRIT, which addresses several computational limitations with prior work by using parallel computing. ESPRIT uses the Needleman–Wunsch algorithm (14) to optimally align each pair of sequences. Through a benchmark study, we demonstrated that global pairwise alignment can provide a much more accurate estimate of microbial richness than multiple alignment. A more important reason, however, is that pairwise alignment allows for parallel computing, while most MSA algorithms can only be used in a single computer. We also developed a new cluster algorithm, referred to as Hcluster within the ESPRIT framework, to handle large-scale clustering problems. Unlike a brute-force method, Hcluster groups sequences into OTUs on-the-fly, while keeping track of linkage information, to overcome memory limitations. By assigning a computational task to hundreds of nodes, ESPRIT is not computationally constrained by the number of sequences to be analyzed, but by the capacity of a computer cluster. Hence, ESPRIT is very suitable for applications such as global ocean survey of microbial populations that may require the analysis of data collected from thousands of locations.
We conducted large-scale experiments to demonstrate the effectiveness of the proposed algorithm. We first performed a simulation study by using a data set generated by sequencing PCR amplicon libraries of known 16S rRNA genes. To our knowledge, this is probably the first benchmark study reported in the literature to assess the estimation accuracy of an algorithm for taxonomy-independent analysis. Our experimental results justified the use of global pairwise alignment for the purpose of estimating microbial community compositions, and showed that the commonly used MUSCLE+DOTUR pipeline may overestimate biodiversity. We then applied ESPRIT to eight seawater samples. The results are very consistent with that of the benchmark study. We finally used ESPRIT to analyze a recently collected air sample consisting of about 350K short reads. It is stated in (15) that microbial communities may be too diversified to be practically tested by amplification and sequencing of 16S rRNA genes. Our results suggest that at least for air samples, it is computationally practical to use the current sequencing technology to conduct 16S rRNA-based biodiversity surveys.
ESPRIT ALGORITHM
The algorithm consists of four modules: (i) removes low-quality reads using various criteria, (ii) computes pairwise distances of reads, (iii) groups reads into OTUs at different dissimilarity levels and (iv) performs statistical inference to estimate species richness. We developed two versions of ESPRIT, one for personal computers (PC) and one for computer clusters (CC). The PC version can process several tens of thousands sequences within a few minutes, while the CC version is able to analyze several hundreds of thousands of reads within one day. The source code and user guide are freely available at http://www.biotech.ufl.edu/people/sun/esprit.html. The readers who have no access to a CC to analyze their data may contact the corresponding author.
Removing low quality reads
The error rate of 454 Life Sciences GS FLX systems is estimated to be five errors per kilobase (http://www.454.com/products-solutions/system-features.asp). However, only a small number of reads account for most of the errors (16), and these outlier reads may be classified at certain dissimilarity levels as rarely occurrent OTUs (i.e. clusters containing only a few members). This will lead to an overestimate of microbial richness in subsequent analyses, since many ecological metrics are computed based on the numbers of rarely occurrent OTUs. To minimize the effects of random sequencing errors, we remove the reads that contain ambiguous nucleotides (N), and those with more than one mismatch with the PCR primer at the beginning of a read. Also, we eliminate the sequences with atypical lengths. The default setting of the algorithm is to retain the reads with a length within 1 SD from the mean length. In order to reduce computational complexity, by following the strategy used in (17), if two sequences are identical or one sequence is a subset of the other, only the longer sequence is retained, and the number of occurrence of each retained sequence is recorded and used in the statistical inference. A similar trimming procedure was also used in (16). ESPRIT allows users to bypass this module to use a sample filtered by a customized trimming procedure.
Computing pairwise distances
We use the Needleman–Wunsch algorithm to optimally align each pair of sequences in a sample, and the quickdist algorithm (4) to compute pairwise distances. More specifically, each pairwise distance equals mismatches, including indels, divided by a sequence length. To avoid overestimating distances between sequences from rapidly diverging variable regions, end gaps are ignored and gaps of any length are treated as a single evolutionary event or mismatch.
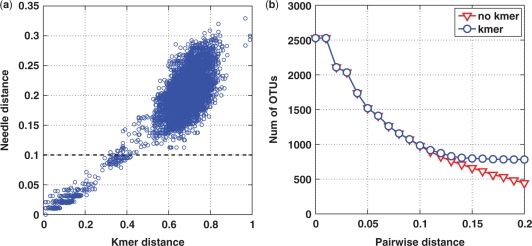
The naive application of the Needleman–Wunsch algorithm to an environmental sample, however, is computationally too expensive. For example, given one full run of 454 data with >400K reads, it is estimated that it would take about 3 years to align all 80 billion pairs of sequences, and need about 1000 GB to store the resulting distance matrix in the PHYLIP format. However, we notice that for the purpose of estimating biodiversity, only the sequence pairs with distances <0.10 are of most interest, which only account for a small fraction of all possible pairs (about 1–5%, Figure 1S). This means possibly 20–100 folds of speedup. Moreover, by removing unwanted pairs, distance information can be stored in a sparse matrix that requires much less memory. In order to rapidly identify the sequence pairs of interest, we compute the k-mer distance of each pair of sequences. We give below a brief description of how it works. A k-mer, also known as k-tuple, is a sub-sequence consisting of k possible nucleotides. By specifying the value of k, a complete alphabet Ω of k-mer is constructed. The number of occurrence of each k-mer in alphabet Ω, also called genomics profile (18,19), is computed for each sequence. Then, the k-mer distance of a pair of sequences is derived as
| 1 |
where f1(i) and f2(i) are the numbers of the occurrence of the i-th k-mer of two sequences, respectively, and L1 and L2 are the lengths of two sequences, respectively. It has been shown that k-mer distances are highly correlated with genetic distances (20). The concept of k-mer counting has been used for various applications, including sequence alignment (10), phylogenetic analysis (21,22) and detection of horizontal gene transfer (23). We used this technique to remove unwanted sequence pairs, and found that by using a k-mer distance of 0.4, most of the sequence pairs of interest can be rapidly identified (Figure 1a). The default setting in ESPRIT is 0.5. Figure 1b shows that removing sequence pairs with k-mer distances larger than the default parameter has a negligible impact on the estimation accuracy for the distance levels <0.1. It should be noted that though the default k-mer threshold works well for all of the 10 data sets tested in this article, it is possible that the parameter may vary for different applications. Hence, we provide an auxiliary program in the software package that allows users to determine the parameter by comparing k-mer distances against genetic distances using a small subset of their samples.
Figure 1.
(a) k-mer distances are highly correlated with genetic distances. (b) Removing sequence pairs with k-mer distances larger than the default threshold has a negligible impact on the estimation accuracy for the distance levels of interest. The experiment was performed on the 53R seawater sample (see Results Section).
In addition to a PC version, we also developed a CC version for calculating both k-mer and genetic distances based on globally aligned sequences. Given one full run of 454 sequences, by using 100 computer nodes, it takes only 3–5 h to finish this module, depending on the availability of computer nodes.
Assigning sequences into OTUs
After a distance matrix is computed, complete-link hierarchical clustering (24) is performed to assign sequences into OTUs of defined sequence variations. Although ESPRIT uses a sparse distance matrix, given a few runs of 454 reads, the size of the matrix can still be quite large. We spent considerable efforts on algorithm development in order to overcome the memory issue, and devised a new clustering algorithm, referred to as Hcluster. The basic idea of the algorithm is to first sort pairwise distances in an ascending order, and then process the distance information on-the-fly. At each step, we classify the clusters being analyzed into ‘active’ or ‘inactive’ clusters. Active clusters are those with known distance information to other clusters, but the information is not enough to decide whether to merge them with other clusters. Inactive clusters, on the other hand, are those with no information at all, or those already merged with other clusters. We only need to maintain the linkage information for active clusters, which is updated at the time when new distance information is processed. We have conducted a large-scale experiment that demonstrated that Hcluster performed very well in the presence of several hundreds of thousands of reads. The accuracy of Hcluster has been benchmarked against DOTUR and MOTHUR. The three methods yielded the exactly same results (Figure 6S). A numerical example is presented in the Supplementary Data to illustrate how Hcluster works.
Clustering is a common problem in bioinformatics. We provide Hcluster as a standalone algorithm in the software package. This algorithm may be useful for other applications where large-scale clustering is needed [e.g. taxonomy-dependent analysis (13)].
Statistical inference of species richness
ESPRIT supports three richness estimators: rarefaction analysis (25), Chao1 (26) and ACE (27,28). Rarefaction allows the calculation of the species richness for a given number of sampled individuals and constructs so-called rarefaction curves. The curve is the number of observed OTUs as a function of the number of sampled sequences. In case of a steep slope, it means that a large fraction of the species diversity is not sampled yet, and more exhaustive sampling will yield a significant number of additional species. Chao1 and ACE are two abundance-based coverage estimators that predict the species richness based on the number of rarely occurrent OTUs. The cluster information generated by ESPRIT allows users to compute other ecological metrics, to derive a consensus sequence of each cluster, and to align the sequences of rarely occurrent OTUs against a database, which may lead to the identification of new organisms.
RESULTS
We conducted large-scale experiments to demonstrate the effectiveness of the newly proposed ESPRIT algorithm. When the PC version of ESPRIT was used, the experiment was performed on a server with eight E5345 Xeon CPUs and 16 GB memory operated on Linux 5.2 system. When the CC version was used, the experiment was performed on a CC administrated by the High-Performance Computing Center at the University of Florida. The detailed computer configuration can be found at http://wiki.hpc.ufl.edu/index.php/Operating_Environment.
Benchmark study
The purpose of the simulation study is 2-fold: (i) to investigate which strategy, pairwise or MSA, is more suitable for taxonomy-independent analysis, and (ii) to assess the estimation accuracy of the ESPRIT algorithm. Although MUSCLE+DOTUR has been used in several metagenomics studies, its performance has never been benchmarked in the literature. This is in part due to the fact that under current technologies, one may never know precisely the ground truth information of the compositions of a microbial community. The benchmark information, however, is critical to evaluate the performance of an algorithm performed on real-world data and to make a meaningful comparison of taxonomic distributions of different environments.
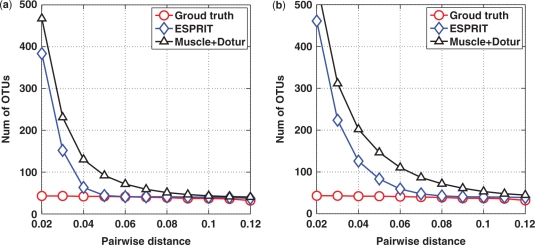
The simulation data consisted of about 340K short sequences, generated by pyrosequencing two PCR amplicon libraries of 43 known 16S rRNA gene fragments using the Roche GS20 system. It was originally used in (16) to study the prebased error rate of the system. We applied both ESPRIT and MUSCLE+DOTUR to the 43 reference gene sequences to estimate the numbers of OTUs defined at various distance levels, also known as lineage-through-time curve in the literature (8). Parameters -maxiters 1 -diags 1 and -sv were used in MUSCLE. The two so-obtained curves served as the ground truth to benchmark the performance of the two algorithms. In order to study how the two algorithms perform in the presence of sequencing errors, we generated two data sets by mapping each read to the 43 reference sequences and retaining the reads that have less than 3% or 5% mismatches with the closest reference sequence. To eliminate statistical variations, each algorithm was run 10 times for each data set. In each run, 10K reads were randomly sampled from each data set. The lineage-through-time curves, averaged over 10 runs, are plotted in Figure 2. Since the ground truth curves generated by ESPRIT and MUSCLE+DOTUR are very similar (Figure 7S presented in the Supplementary Data), for ease of presentation, only the curve of ESPRIT is plotted. From the figure, we observe that while both algorithms always lead to an overestimate of the number of OTUs due to the presence of sequencing errors, pairwise alignment can provide a more accurate estimate of microbial richness than multiple alignment. For example, for the first data set, at the 0.05 distance level, the numbers of OTUs estimated by ESPRIT and MUSCLE+DOTUR are 44 (95% CI: 43–45) and 92 (95% CI: 82–102), respectively, while the ground truth is 42 (Figure 2a).
Figure 2.
Lineage-through-time curves generated by using ESPRIT and MUSCLE+DOTUR algorithms performed on simulation data with each read containing up to (a) 3% and (b) 5% sequencing errors.
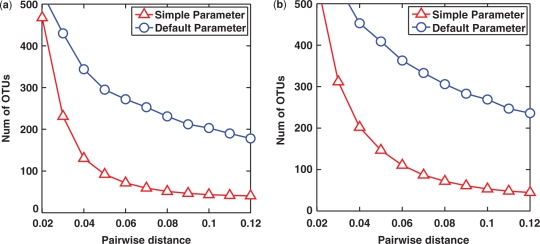
One may wonder whether the disparity between the results of the two methods is due to the compromise on the parameter settings of MUSCLE. To answer this question, we repeated the experiment by using the default parameter of MUSCLE (-maxiters 16) (29). The results are reported in Figure 3. Though computationally expensive, using the default parameter results in a much better alignment in terms of the sum of pairwise alignment scores. Interestingly, we observe that the estimate obtained by using the default parameter is much worse than that obtained by using the simple parameter. This can be explained by the fact that MSA aims to minimize the sum of pairwise alignment scores, ignoring the fact that a large proportion of sequence pairs originate from distantly related OTUs. Since many existing methods used MSA for taxonomy-independent analysis, this issue merits further investigation, which is presented in the Supplementary Data (Section 1S). We also conducted a benchmark study comparing the prediction performance of ESPRIT with those of NAST (30) and RDP-Pyro (13) in Section 2S.
Figure 3.
Lineage-through-time curves generated by MUSCLE+DOTUR using simple or default parameters performed on simulation data with each read containing up to (a) 3% and (b) 5% sequencing errors.
Experiments on eight seawater samples
We applied ESPRIT to reanalyze eight seawater samples downloaded from (4). The DNA materials within environmental samples were collected from eight different locations in the Atlantic and Pacific Oceans, respectively, as a part of efforts to develop a global description of microbial diversities in the ocean. A total of 118 000 PCR amplicons were sequenced that covered the V6 hyperviable region of rRNAs. The 454 reads have undergone a systematic trimming process. Thus, we bypassed the trimming procedure. The number of sequences for each sample ranges from 5000–17 666 (see Table 1). The interested reader may refer to (4) for a detailed description of the data and sample preparations.
Table 1.
Running time of the PC version of ESPRIT performed on eight seawater samples
| Data sets |
||||||||
|---|---|---|---|---|---|---|---|---|
| 112R | 115R | 137 | 138 | 53R | 55R | FS312 | FS396 | |
| Number of reads |
9282 |
11 005 |
13 097 |
14 374 |
5000 |
13 902 |
4835 |
17 666 |
| CPU time | 2 m 54 s | 4 m 13 s | 4 m 28 s | 6 m 19 s | 59 s | 7 m 31 s | 49 s | 6 m 34 s |
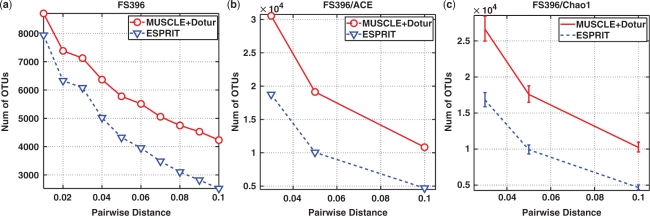
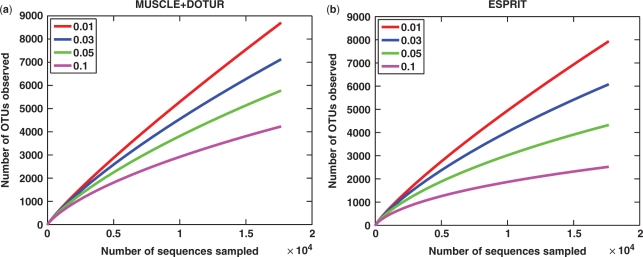
We reran MUSCLE+DOTUR by using the parameters provided by (4). The default parameters of ESPRIT were used. Due to space limitations, only the results of sample FS396 are presented in the main text. FS396 contains 17 666 reads after trimming, and is the largest data set in size among the eight samples. However, the results of the other seven seawater samples, presented in the Supplementary Data (Figures S8 and S9), are very consistent with that of FS396. The lineage-through-time curves, ACE and Chao1 estimates at three different distance levels are depicted in Figure 4. We observe that ESPRIT gives a much lower estimate of species richness than the competing method, which is consistent with the result of the simulation study described above. For example, at the distance of 0.05, the number of observed OTUs and the ACE and Chao1 estimates obtained by ESPRIT are only about 50–70% of those obtained by MUSCLE+DOTUR. This becomes more evident in Figure 5, where the results of the rarefaction analysis of FS396 are presented. Though the existing pipeline overestimates the diversity of microbes, the conclusion of (4) still holds that the microbial diversity of seawater samples is much larger than previously reported, and the steep slopes of the rarefaction curves, even at relatively large genetic distances (e.g. 0.1), suggest that a large fraction of species have not been sampled yet. We notice that the disparity between the results of the two methods is much larger than that in the simulation study (Figures 2, 4, S8 and S9). One possible reason is that the sequences in the seawater samples are more diverse than those in the simulation data (see Section 1S for a detailed discussion).
Figure 4.
(a) Lineage-through-time curves, (b) ACE and (c) Chao1 estimates generated by using ESPRIT and MUSCLE+DOTUR algorithms performed on the FS396 data. Error bars of Chao1 estimates represent the 95% confidence interval.
Figure 5.
Rarefaction curves generated by using (a) MUSCLE+DOTUR and (b) ESPRIT performed on the FS396 data.
ESPRIT is computationally very efficient. As can be seen from Table 1 where the CPU times of the PC version of ESPRIT performed on the eight seawater samples are recorded, ESPRIT can process several tens of thousands of sequences within a few minutes. As both the results of (4) and our reanalysis of their samples suggest deeper sequencing is required for accurate estimation of microbial richness of environmental samples, the CC version of ESPRIT is expected to be computationally more efficient than the PC version, as we will see in the next section.
Experiments on an air sample
We applied ESPRIT to an air sample recently collected from Iraq. The small subunit rRNA gene fragments that cover the V6 hyperviable region of rRNAs were amplified, cloned and pyrosequenced using 454 Life Sciences systems. A total of 348 952 sequences of an average of 275 nt in length were obtained. This is one of the largest 16s rRNA-based biodiversity surveys conducted using air samples. The number of sequences is one order of magnitude larger than those of the seawater samples we considered in the previous section. Interested reader may refer to the Supplementary Data for a detailed description on how the air sample was prepared.
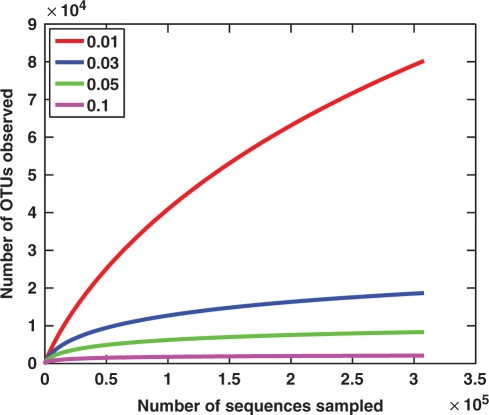
The CC version of ESPRIT with the default parameters was used. After the trimming procedure, 40 928 (about 13%) reads were removed as low-quality reads. Figure 6 presents the results of the rarefaction analysis of the OTUs defined at four different sequence variations, ranging from 0.01 to 0.1. We observe that, in contrast with the results of the seawater samples, the rarefaction curves saturate even at a relatively small genetic distance of 0.03, indicating that additional sampling may not lead to significantly increased estimates of total species diversity. The results of ACE and Chao1 estimators are reported in Table 2. At the distance levels of 0.05 and 0.1, the numbers of observed OTUs are already very close to the ACE and Chao1 estimates. These experimental results suggest that, at least for air samples, the current sequencing technology is sufficient to conduct 16S rRNA-based biodiversity surveys. The experiment was performed on a small CC consisting of 100 nodes, and it took about 10 h to finish the entire analysis.
Figure 6.
Rarefaction analysis of an air sample. Rarefaction curves are shown for OTUs with sequence variations that do not exceed 1, 3, 5 or 10%.
Table 2.
The number of observed OTUs, the ACE and Chao1 estimates of an air sample at four different distance levels
| Pairwise distance |
||||
|---|---|---|---|---|
| 0.01 | 0.03 | 0.05 | 0.1 | |
| OTUs | 80 238 | 18 686 | 8344 | 2109 |
| ACE |
147 266 |
23 894 |
9664 |
2262 |
| Chao1 | 138 376 | 23 921 | 9748 | 2293 |
| Upper | 139 911 | 24 302 | 9932 | 2362 |
| Lower | 136 881 | 23 566 | 9585 | 2242 |
The 95% CIs of the Chao1 estimates are also provided.
DISCUSSIONS
Equipped with next-generation sequencing technology, researchers now start to sequence many millions of sequences for applications such as global ocean surveys and epidemiological studies with many patients. Parallel computing that distributes computation to hundreds or even thousands of nodes seems at this time to be the only viable approach. We have demonstrated that the newly proposed ESPRIT algorithm can process several hundreds of thousands of sequences within 10 h by using a relatively small CC. To our knowledge, no existing methods can efficiently handle such large data. The two key components of ESPRIT are the use of pairwise instead of multiple sequence alignment, which allows for parallel computing, and the development of Hcluster that performs hierarchical clustering using online learning to address the memory issue. We have demonstrated that MSA is not only computationally expensive, but also tends to overestimate microbial diversity (see Section 1S). In order to process many millions of reads, researchers may need to collaborate with a supercomputing center (e.g. IBM Roadrunner which has about 20 000 nodes). Our ESPRIT algorithm can easily be modified to work on any cluster. The computational complexity of ESPRIT is quadratic with respect to the number of reads. At Roadrunner's full capacity, we estimate that it will take 1 s, 10 h and 40 h to process 1, 50 and 100 million reads, respectively. Of course, this is an overoptimistic estimation, since it focuses only on computational complexity and assumes that the resources of the entire supercomputing center are available for this singular task. ESPRIT, however, does provide a promising direction for analyzing large collection of 16S rRNA data. We will continue to optimize the performance of ESPRIT and release the software to the community. We believe that ESPRIT will evolve to be a powerful tool for metagenomics study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the editor and three reviewers for their valuable comments to improve the quality of the article. The simulation study presented in Section 1S was inspired by one of the reviewers' comment.
Conflict of interest statement. None declared.
REFERENCES
- 1.Eisen JA. Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol. 2007;5:e82. doi: 10.1371/journal.pbio.0050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothberg JM, Leamon JH. The development and impact of 454 sequencing. Nat. Biotechnol. 2008;26:1117–1124. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- 3.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braver man MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl Acad. Sci. USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keijser B, Zaura E, Huse SM, van der Vossen J, Schuren F, Montijn RC, ten Cate JM, Crielaard W. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 6.Borneman J, Triplett EW. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sait M, Hugenholtz P, Janssen PH. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 2002;4:654–666. doi: 10.1046/j.1462-2920.2002.00352.x. [DOI] [PubMed] [Google Scholar]
- 8.Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y, Breitbart M, McNairnie P, Rohwer F. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics. 2006;7:57. doi: 10.1186/1471-2105-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 15.Gans J, Woilinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 16.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber JA, Welch DBM, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 18.Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 19.Karlin S, Mrazek J, Campbell A. Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 1997;179:3899–3913. doi: 10.1128/jb.179.12.3899-3913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. Local homology recognition and distance measures in linear time using compressed amino acid alphabets. Nucleic Acids Res. 2004;32:380–385. doi: 10.1093/nar/gkh180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavromatis K, Ivanova N, Barry K, Shapiro H, Goltsman E, McHardy AC, Rigoutsos I, Salamov A, Korzeniewski F, Land M, et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat. Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Yu F, Liu L, Farmerie W. Estimating microbial population densities based on genomic signatures. Proc. Intl. Conf. Bioinform. Comput. Biol. 2007;1:163–168. [Google Scholar]
- 23.Dalevi D, Dubhashi D, Hermansson M. Bayesian classifiers for detecting HGT using xed and variable order markov models of genomic signatures. Bioinformatics. 2006;22:517–522. doi: 10.1093/bioinformatics/btk029. [DOI] [PubMed] [Google Scholar]
- 24.Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd. New York: Wiley; 2001. [Google Scholar]
- 25.Hurlbert SH. The non-concept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 26.Chao A. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- 27.Chao A, Lee SM. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 1992;87:210–217. [Google Scholar]
- 28.Chao A, Ma MC, Yang MCK. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika. 1993;80:193–201. [Google Scholar]
- 29.Edgar RC. MUSCLE user guide. Technical Report. 2004 Available at http://www.drive5.com/muscle/docs.htm. [Google Scholar]
- 30.DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.