Abstract
Many longitudinal studies of aging collect genetic information only for a sub-sample of participants of the study. These data also do not include recent findings, new ideas and methodological concepts developed by distinct groups of researchers. The formal statistical analyses of genetic data ignore this additional information and therefore cannot utilize the entire research potential of the data. In this paper, we present a stochastic model for studying such longitudinal data in joint analyses of genetic and non-genetic sub-samples. The model incorporates several major concepts of aging known to date and usually studied independently. These include age-specific physiological norms, allostasis and allostatic load, stochasticity, and decline in stress resistance and adaptive capacity with age. The approach allows for studying all these concepts in their mutual connection, even if respective mechanisms are not directly measured in data (which is typical for longitudinal data available to date). The model takes into account dependence of longitudinal indices and hazard rates on genetic markers and permits evaluation of all these characteristics for carriers of different alleles (genotypes) to address questions concerning genetic influence on aging-related characteristics. The method is based on extracting genetic information from the entire sample of longitudinal data consisting of genetic and non-genetic sub-samples. Thus it results in a substantial increase in the accuracy of statistical estimates of genetic parameters compared to methods that use only information from a genetic sub-sample. Such an increase is achieved without collecting additional genetic data. Simulation studies illustrate the increase in the accuracy in different scenarios for datasets structurally similar to the Framingham Heart Study. Possible applications of the model and its further generalizations are discussed.
Keywords: stochastic process model, allostatic load, age-dependent physiological norm, adaptive capacity, stress resistance
1. Introduction
The influence of genes on aging, health and longevity is mediated by thousands of biological and physiological variables which are also affected by environmental, behavioral and other factors. Some of such variables are measured in longitudinal studies of aging, health and longevity. That is why the data on genetic markers collected for participants of a longitudinal study are probably most appropriate for evaluating the genetic contribution to the aging-related decline in the health/well-being status and the life span. Such data, however, often cannot be collected for all participants of the study. This is because: (i) the large-scale collection of genetic data is a relatively new business, thus, some individuals, who initially participated in a longitudinal study, have already died or dropped out of a population; (ii) obtaining genetic information is still an expensive business and cannot be performed at the same scale as medical examinations or a sociological survey; (iii) not all individuals who agreed to participate in a medical examination or to respond to the survey’s questionnaire agree to participate in a genetic analysis. Thus, the presence of genetic information divide participants of a longitudinal study into two groups: one (the genetic group) includes those for whom genetic data were also collected. The other (the non-genetic group) consists of those for whom longitudinal data are available but genetic information was not collected.
Such a situation when information on covariates essential for analyses of risks is missing for some sub-sample of individuals (either due to cost limitations or by the study design) is typical in epidemiological studies. For example, two-stage designs are routinely used in epidemiology when a disease status (or other general information) is ascertained for a large group of individuals at the first stage and information on covariates essential for analyses of their relation to the risk of the disease is collected at the second stage for smaller sub-samples of individuals. Statistical methods for analyses of such data are well developed for regression models (Breslow and Cain, 1988; Breslow and Holubkov, 1997a; Breslow and Holubkov, 1997b; Cain and Breslow, 1988; Scott and Wild, 2001; Scott et al., 2007). One of the main advantages of such methods is that they use information from the first and second stages to estimate regression parameters. This can lead to a considerable improvement in the efficiency of estimates compared to the estimates based on the second stage data alone. Applications of such designs and methods in genetic epidemiology are also discussed in the literature (e.g., Bureau et al., 2008; Chatterjee and Chen, 2007).
A traditional way of evaluating effects of genes on individuals’ health/well-being/survival status is to directly estimate respective hazards (e.g., incidence or mortality rate) for carriers of a selected allele (genotype). Such practice is completely justified in the absence of data about other factors and processes affecting these characteristics. The advantage of longitudinal data for the genetic studies of aging and longevity is in the opportunity to estimate not only direct genetic effects on morbidity and mortality but also indirect genetic effects mediated by age trajectories of physiological variables collected in the longitudinal study (which may modulate mechanisms of aging not directly measured in longitudinal data).
The purpose of this paper is to elaborate a genetic model for studying longitudinal data on aging, health, and longevity which would permit: 1) joint analyses of genetic and non-genetic data to make use of all available information and increase the accuracy of estimates compared to analyses of genetic data alone; 2) evaluation of indirect genetic effects mediated by age trajectories of physiological variables collected in a longitudinal study; and 3) incorporation of essential mechanisms of aging-related changes in organisms that are not directly measured in longitudinal data but can be estimated from individual age trajectories of physiological indices and data on mortality or morbidity. The stochastic process model (SPM) of human mortality and aging (Manton and Yashin, 2000; Woodbury and Manton, 1977; Yashin, 1985; Yashin and Manton, 1997) is the conceptual approach in this study and its extension presented in this paper has all three above-mentioned properties. The important feature of the SPM is a biologically-justified U- or J- shaped risks as functions of respective indices. Such shapes of the risk functions are observed for different physiological indices (Allison et al., 1997; Boutitie et al., 2002; Kulminski et al., 2008; Kuzuya et al., 2008; Mazza et al., 2007; Okumiya et al., 1999; Protogerou et al., 2007; Troiano et al., 1996; Witteman et al., 1994). The original SPM was recently modified (Yashin et al., 2007b) to include major concepts of aging known to date: age-specific physiological norms (Lewington et al., 2002; Palatini, 1999; Westin and Heath, 2005), allostasis and allostatic load (Karlamangla et al., 2006; Seeman et al., 2001), the decline in adaptive capacity with age (homeostenosis) (Lund et al., 2002; Troncale, 1996), the decline in stress resistance with age (Hall et al., 2000; Ukraintseva and Yashin, 2003; Yashin et al., 2006), and stochasticity (Goldberger et al., 2002). The one- and two-dimensional versions of the model were successfully applied to different data sets to reveal complicated interplay among different components of aging-related changes in humans (Yashin et al., 2007c; Yashin et al., 2007d; Yashin et al., 2008a). The model presented in section 2 of this paper is a step forward in analyzing contribution of genes to dynamic regularities in aging-related changes in a human organism. This model incorporates information on genetic markers collected for a sub-sample of participants of a longitudinal study and permits evaluation of all above-mentioned characteristics (age-specific norms, decline in stress resistance, etc.), as well as respective hazard rates, for carriers and non-carriers of a selected allele (genotype) to address questions concerning genetic influence on these aging-related characteristics (here we formulated the model for two types of individuals: carriers and non-carriers of some selected allele/genotype, however, its extension to the case of many alleles/genotypes is straightforward). The method is based on extracting genetic information from the entire sample of longitudinal data consisting of genetic (those with available genetic information) and non-genetic (those for whom genetic information was not collected) sub-samples. The group of individuals with genetic data becomes automatically divided into subgroups of carriers and non-carriers of respective alleles or genotypes. The non-genetic group consists of carriers of the same genotypes identified in the genetic group and, hence, non-genetic data contain information about genetic influence on all phenotypes observed in a longitudinal study. We develop statistical methods for extracting genetic information from the entire sample of longitudinal data consisting of genetic and non-genetic sub-samples. This joint analysis results in a substantial increase in the accuracy of statistical estimates of genetic parameters (without collecting additional genetic data) compared to methods that use only information from a genetic sub-sample. Simulation studies illustrating the increase in the accuracy in different scenarios for datasets structurally similar to the Framingham Heart Study (FHS) (Dawber et al., 1951) are presented in section 3 and in Supplementary Material. The last section summarizes the results and discusses perspectives of further research in this area.
2. SPM for joint analysis of genetic and non-genetic data from longitudinal studies
2.1. General model
Yashin et al. (2007b) suggested the stochastic model that includes several major concepts of aging known to date and that links individual trajectories of physiological or other indices measured in longitudinal data and mortality or morbidity risks. The model uses several assumptions in description of the dynamic properties of physiological indices and the function of the mortality/morbidity risk. It is assumed that the age dynamics of physiological indices is modeled by a multidimensional stochastic process (with a normally distributed initial value). This process represents two main components of the age dynamics. The first one corresponds to the basic regularities of the age-related physiological changes and the second is a stochastic component summarizing the effects of external and internal disturbances in the dynamics of the indices. The basic regularities include the notion of allostatic adaptation, i.e., the average trajectories of physiological indices which the organisms are forced to follow and which represent average effects of interplay among factors controlled by the ontogenetic program, senescence, and long-acting environmental stresses exceeding the limits of the homeostatic regulation in human organisms. The equation also includes a feedback mechanism which represents the homeostatic regulation and forces the trajectories of physiological indices to return to their average levels in case of disturbances (deviations from these levels). The mortality/morbidity risk is assumed as a quadratic hazard function to capture J- or U-shapes of the risks considered as a function of risk factors (physiological indices). The notion of physiological norms of indices is introduced to represent the values of indices with minimal mortality/morbidity risks at respective ages. Deviations from this norm elevate the mortality/morbidity risk (compared to the baseline level) and it is assumed that the magnitude of this elevation is age-dependent (i.e., the magnitude of the U-shape of the risk function changes with age) representing the decline in stress resistance with age.
In this paper, we extend the Yashin et al. (2007b) model including the dependence of the dynamics of physiological indices and hazard rates on genetic markers. The description corresponds to an assumption that a population under study (e.g., participants of a longitudinal study) is a mixture of carriers and non-carriers of some selected allele (or genotype) with initial proportions p and 1 − p, respectively. We assume that for some portion of this population genetic data are collected. Availability of such data allows one to hypothesize that longitudinal data for carriers and non-carriers of the selected allele (genotype) are represented by the same model with possibly different (allele- or genotype-specific) parameters describing the evolution of physiological indices and the shape of the mortality/morbidity rate.
Let a discrete random variable Z (Z = 0, 1; P(Z = 1) = p) characterize the absence (Z = 0) or presence (Z = 1) of a selected allele (or genotype) in the genome of an individual randomly selected from a population. Let Yt be a continuously changing random covariate (a vector of physiological indices). We assume that the evolution of Yt depends on the presence (or absence) of a selected allele (genotype) in the genome and it may be described by the following stochastic differential equation with coefficients depending on Z:
| (1) |
Here Yt (t is age; we omit dependence of the process on Z for conciseness) is a k-dimensional stochastic process with the initial condition Yt0. We assume that the conditional distribution of Yt0 given Z (p(Yt0 |Z = z), z = 0, 1) is normal with mean m (z,t0) = mz,0 and variance γ(z,t0) = γz,0. Wt is a (k-dimensional) vector Wiener process independent of Yt0 and Z. It describes external disturbances affecting these covariates and incorporates stochasticity into the model. The strength of disturbances is characterized by the matrix of diffusion coefficients B(Z, t). The vector-function f1(Z, t) (having the same dimension as Yt) introduces the notion of allostasis into the model. It describes the age trajectory of a physiological state which organisms are forced to follow by the process of allostatic adaptation (McEwen and Wingfield, 2003). This function describes average trajectories of physiological indices resulting from a complicated interplay among factors controlled by the ontogenetic program, senescence, and long-acting environmental stresses in human organisms and may be referred to as the “mean allostatic state.” Dependence of this function on Z indicates that mechanisms of allostatic adaptation may differ for groups of individuals characterized by different values of Z (i.e., in carriers and non-carriers of a selected allele/genotype). The matrix a(Z, t) describes the mechanism of decline in adaptive (homeostatic) capacity in an aging organism (see example in section 2.1 of Yashin et al., 2007b). The elements of this matrix correspond to the rate of adaptive response to any deviation of physiological indices Yt from f1(Z, t) (i.e., the homeostatic adaptation of physiological indices Yt to the allostatically prescribed trajectories f1(Z, t)). Dependence of this matrix on age captures average age-related changes in the “homeostatic capacity” of a human organism. Its dependence on Z captures potential differences in adaptive capacity in carriers and non-carriers of a selected allele/genotype.
Let the mortality rate conditional on Yt and Z be:
| (2) |
Here the scalar function μ0(Z,t) is the background (baseline) hazard characterizing the residual mortality rate, which would remain if all covariates Yt follow their optimal trajectories, i.e., coincide with the vector-function f(Z, t). Thus, μ0(Z,t) is associated with death from factors other than those involved in the quadratic term and represented by Yt (i.e., with unmeasured factors). Its dependence on Z indicates the possibility that the effect of unobserved factors (which may be of genetic or non-genetic origin) on the mortality risk is different in carriers and non-carriers of a selected allele/genotype. The function f(Z, t) is introduced to explicitly associate changes in the “optimal” physiological state with the minimum of hazard at respective ages. It has a meaning of the age-specific physiological norm corresponding to a minimum risk of death at specific ages. It may differ from f1(Z, t) since the process of allostatic adaptation (considered as an organism’s response to persistent disturbances) does not necessarily result in the optimal physiological state. Thus, the difference between f1(Z, t) and f(Z, t) provides the measure of the allostatic load. Dependence of f(Z, t) on Z indicates the possibility that carriers and non-carriers of a specific allele/genotype may have different age-trajectories of physiological norms. Q(Z, t) is a non-negative-definite symmetric matrix (for all values of Z and t) of respective dimension (k × k). Its dependence on age is assumed to allow for capturing the decline in stress resistance. For example, in a one-dimensional case, an increasing pattern of Q(Z, t) with age (t) indicates that the branches of respective U-shaped risk function are getting steeper with age. This means that the range of “acceptable” deviations of the respective risk factor (represented by Yt) from its “optimal” values (represented by f (Z, t)), which result in a moderate increase in the risk of death, is getting narrower with age. This, in turn, is an indicator of decline in stress resistance with age. Dependence of Q(Z, t) on Z indicates the possibility that carriers and non-carriers of a specific allele/genotype differ with respect to the aging-related decline in stress resistance.
2.2. Likelihood function for data from genetic group
Let the sequence , τi represent the results of ni + 1 measurements of the process Yt at ages , j = 0…ni, and the life span (which may be censored) related to ith individual from the genetic group (i.e., for whom information on Z is known). The following likelihood function can be used to estimate the model parameters for is the number of individuals with Z = z, z = 0, 1) individuals from this group:
| (3) |
The products in (3) are calculated over individuals with respective value of z, z = 0, 1, and the likelihood for ith individual with Z = z is
| (4) |
The hazard rate at age t for ith individual with Z = z, μ̄i(z,t), is given by
| (5) |
Functions mi (z,t) and γi(z,t) in (4) and (5) are mean and variance of the conditional distribution P(Yt ≤ y|Z = z, T > t), which satisfy the following ordinary differential equations:
| (6) |
| (7) |
at the intervals between the observation times, , with initial conditions , and γz,0, 0, …, 0, respectively. Thus, the trajectories of mi (z,t)and γi(z,t) defined by equations (6) and (7) differ for different individuals. Consequently, the estimates of the chances of death for individuals having different observed values of the respective covariates will also differ. δi denotes a censoring indicator (1 for died, 0 for censored), is the age of the latest measurement of the physiological index before death/censoring at τi, and is the determinant of the matrix , z = 0,1.
2.3. Likelihood function for data from non-genetic group
The non-genetic group is a discrete mixture of carriers and non-carriers of alleles or genotypes measured in the first (genetic) group. If the genetic subgroup has been randomly selected from the data, then the proportions of carriers and non-carriers of respective allele (genotype) in genetic and non-genetic groups are about the same. The likelihood function of longitudinal data for the non-genetic group is a function constructed for such a heterogeneous population. Let Nng (t0) be the number of individuals in the non-genetic group. Since the genotypes of respective individuals are unknown, the likelihood function of these data is
| (8) |
where and are calculated for ith individual from the non-genetic group using (4).
2.4. Joint analysis of genetic and non-genetic data
One can see that, although the likelihood functions constructed for genetic and non-genetic data have different structures, they depend on the same parameters (those of functions μ0 (Z,t), Q(Z, t), f(Z, t), f1(Z, t), a(Z, t), and B(Z, t)). This property suggests that the joint analysis of such data will improve the accuracy of parameter estimates compared to the analysis of data from the genetic group alone. The likelihood function for genetic and non-genetic data is the product of the likelihoods constructed for genetic and non-genetic groups:
| (9) |
where Lg and Lng are given by (3) and (8). Maximizing this likelihood, we will obtain the parameter estimates that characterize the dynamics of the stochastic process Yt describing the trajectories of physiological indices and the mortality rates for carriers and non-carriers of selected allele (genotype). One can also test the hypotheses on differences in respective parameters in carriers and non-carriers of allele (genotype) estimating the model with equal parameters for carriers and non-carriers (i.e., some or all functions from the above list do not depend on Z) and the general model with parameters depending on Z, and comparing these two models using the likelihood ratio test (see examples in Simulation studies S1 in Supplementary Material). If such differences are significant, this will indicate the presence of a genetic effect in respective component of the model (e.g., in age-specific norms). If they are not, then the respective component can be well modeled by a general “population” function (i.e., not depending on Z).
3. Results of simulation study: Comparison of accuracy of estimates in joint analysis of genetic and non-genetic data and in analysis of genetic data alone
We performed a simulation study to check performance of the model in a one-dimensional case and compare the accuracy of estimates in the joint analysis of genetic and non-genetic data and in the analysis of genetic data alone. In computer simulations, we used a discrete-time version of the general model (1)–(2). We assumed that the background mortality μ0(Z,t) in (2) is the Gompertz hazard μ0(Z,t) = aμ0 (Z)ebμ0(Z)(t−tmin), where tmin = 30. The quadratic hazard terms, Q(Z, t), the mean allostatic state, f1(Z, t), and the age-dependent norms, f(Z, t), are taken as linear functions of age: Q(Z,t) = aQ (Z) + bQ (Z)t, f1(Z, t) = af1 (Z) + bf1 (Z)(t − tmin), and f(Z, t) = af (Z) + bf (Z)(t − tmin). The rates of adaptive regulation, a(Z, t), and the diffusion coefficients, B(Z, t), are assumed constant: a(Z,t) = aY (Z), and B(Z,t) = σ1(Z). The initial distribution of Yt0 is normal with the mean f1 (Z,t0) and the variance . The initial proportion of a hypothetical allele (or genotype) in a population (i.e., P(Z = 1)) is denoted by p. Parameters to be estimated in this model are: ln aμ0 (Z) (note that we estimated ln aμ0 (Z) instead of aμ0 (Z) because the values of aμ0 (Z) in this study are close to zero, which is typical for human data and close to the boundary of acceptable values for this parameter), bμ0 (Z), aQ (Z), bQ (Z), aY (Z), σ0 (Z), σ1(Z), af1 (Z), bf1 (Z), af (Z), bf (Z), for Z = 0, 1, and p. Ages at entry into the study were simulated as a discrete random variable uniformly distributed over the interval from 30 to 60. The interval between observations of Yt equals two years. The number of observations (exams) is 25. This structure resembles the Framingham Health Study (FHS) data (Dawber et al., 1951). We simulated 100 data sets, with 2500 individuals in each sample (which is roughly comparable with the sex-specific sample sizes in the FHS). We assigned the values of Z (1 and 0) to each individual in the sample with probabilities p and 1 − p and simulated the age-trajectories of the process Yt (a hypothetical physiological index) and probabilities of death at discrete time intervals (given by (1) and (2), respectively) using the generator of random numbers implemented in MATLAB.
The likelihood maximization was performed using the constrained optimization procedure of MATLAB’s optimization toolbox (MathWorks Inc., 2008). Restrictions on acceptable ranges of parameter values are necessary for functions in the mortality risk (2) and the stochastic equation for the risk factor Yt (1). These restrictions reasonably impose constraints on parameters of: a) an initial distribution Yt0 (to ensure a negligible probability of values outside the “acceptable range” for the process Yt, which was arbitrarily taken as the interval from 10 to 100); b) functions f1(Z, t) and f(Z, t) (to ensure that their values are within the acceptable range for the process Yt for ages 30 to 105); c) the rate of adaptive regulation a(Z, t) (to ensure that the feedback coefficient in (1) does not become too small or positive and that the trajectories of Yt tend to f1(Z, t)); d) the background hazard μ0 (Z,t) (to ensure non-negative values for each age, a non-decreasing age pattern, and trajectories of the hazard rates typical of human data); and e) the quadratic hazard terms Q(Z, t) (to ensure non-negative values for each age from 30 to 105).
To evaluate an increase in the accuracy of the estimates in the joint modeling compared to the methods using data from the genetic sample alone, we used three models. First, we assumed that genetic information is available only for a subset of 500 individuals and maximized the likelihood Lg in (3) using only data on 500 individuals. Second, we assumed that genetic information is available for the entire sample. In this case, the likelihood function Lg in (3) was maximized using data on all 2500 individuals. Third, we used the joint model (with the likelihood L in (9)) assuming that genetic information is available only for a subset of 500 individuals (with respective proportions of carriers (p) and non-carriers (1 − p) of a hypothetical allele or genotype) and the remaining 2000 individuals in a sample constituting the non-genetic group (for which information on the genotype is unknown). The results of this simulation study are shown in Table 1 and Figs. 1–2.
Table 1.
Simulation study: Means (standard deviations) of parameter estimates for carriers (Z = 1) and non-carriers (Z = 0) of a hypothetical allele or genotype in 100 simulated data sets in case of three methods of parameter estimates: 1) Analyses of genetic data when only a sub-sample of 500 individuals contains genetic information (“Genetic (500)”); 2) Analyses of genetic data when the entire sample (2500 individuals) contains genetic information (“Genetic (2500)”); and 3) Joint analyses of genetic (500 individuals) and non-genetic (2000 individuals) data (“Joint (500+2000)”).
| ln a μ0 | b μ0 | a Q ·104 | b Q ·105 | a Y | σ0 | σ1 | a f1 | b f1 | a f | b f | p | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic (500) | ||||||||||||
| Z = 1: | −6.92 (2.347) | 0.099 (0.051) | 0.03 (1.435) | 0.20 (0.280) | −0.20 (0.010) | 6.04 (0.417) | 4.99 (0.101) | 39.86 (0.764) | 0.51 (0.034) | 36.69 (7.249) | 0.00 (0.287) | |
| Z = 0: | −8.65 (1.967) | 0.110 (0.039) | 0.18 (0.664) | 0.12 (0.122) | −0.20 (0.006) | 5.95 (0.198) | 4.00 (0.043) | 45.00 (0.351) | 0.50 (0.012) | 49.85 (3.568) | −0.21 (0.168) | |
| Genetic (2500) | ||||||||||||
| Z = 1: | −6.10 (0.353) | 0.082 (0.010) | 0.28 (0.855) | 0.15 (0.157) | −0.20 (0.005) | 6.02 (0.162) | 5.00 (0.045) | 39.97 (0.347) | 0.50 (0.016) | 38.83 (3.567) | −0.06 (0.163) | |
| Z = 0: | −8.08 (0.454) | 0.101 (0.010) | 0.29 (0.271) | 0.10 (0.050) | −0.20 (0.002) | 6.00 (0.093) | 4.00 (0.018) | 44.99 (0.151) | 0.50 (0.006) | 50.06 (1.272) | −0.21 (0.072) | |
| Joint (500+2000) | ||||||||||||
| Z = 1: | −6.12 (0.575) | 0.082 (0.016) | 0.39 (0.950) | 0.12 (0.175) | −0.20 (0.006) | 6.03 (0.227) | 5.00 (0.055) | 39.93 (0.429) | 0.50 (0.020) | 39.85 (4.012) | −0.10 (0.184) | 0.252 (0.010) |
| Z = 0: | −8.19 (0.798) | 0.103 (0.014) | 0.24 (0.372) | 0.11 (0.065) | −0.20 (0.003) | 5.99 (0.101) | 4.00 (0.019) | 45.01 (0.169) | 0.50 (0.006) | 49.72 (2.055) | −0.19 (0.094) | |
| True values | ||||||||||||
| Z = 1: | −6.0 | 0.08 | 0.5 | 0.1 | −0.2 | 6.0 | 5.0 | 40.0 | 0.5 | 40.0 | −0.1 | 0.25 |
| Z = 0: | −8.0 | 0.1 | 0.3 | 0.1 | −0.2 | 6.0 | 4.0 | 45.0 | 0.5 | 50.0 | −0.2 | |
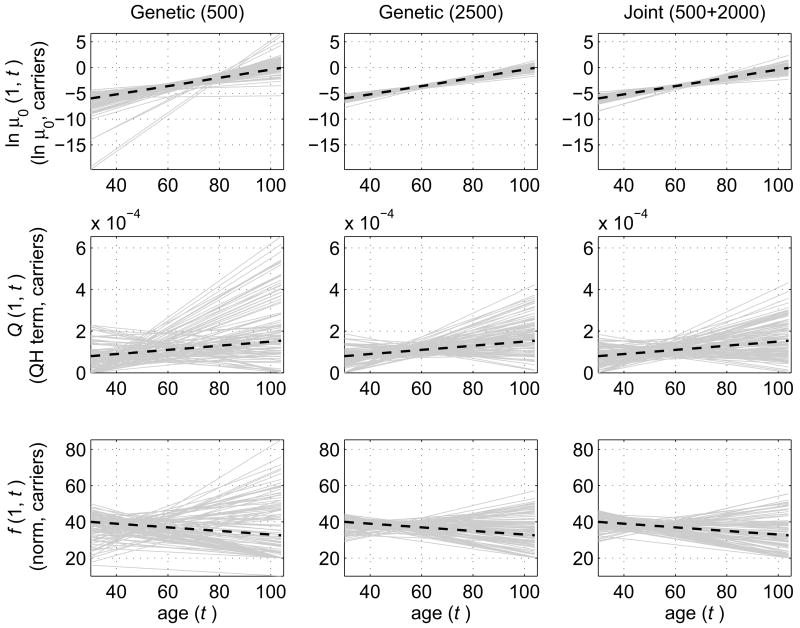
Fig. 1.
Simulation study: Comparison of estimates for carriers of a hypothetical allele (genotype) obtained in 100 simulated data sets by three methods. Left column: Estimates of age trajectories (solid grey lines) of logarithm of baseline hazard (ln μ0 (1,t)), quadratic hazard (QH) terms (Q(1, t)), and age-dependent norms (f(1, t)) calculated using only sub-samples with genetic information (500 individuals). Middle column: Similar estimates when the entire sample (2500 individuals) contains genetic information. Right column: The estimates calculated using the joint analysis of genetic (500 individuals) and non-genetic (2000 individuals) data. Respective “true” trajectories used for simulation of data are shown as dashed black lines; t denotes age.
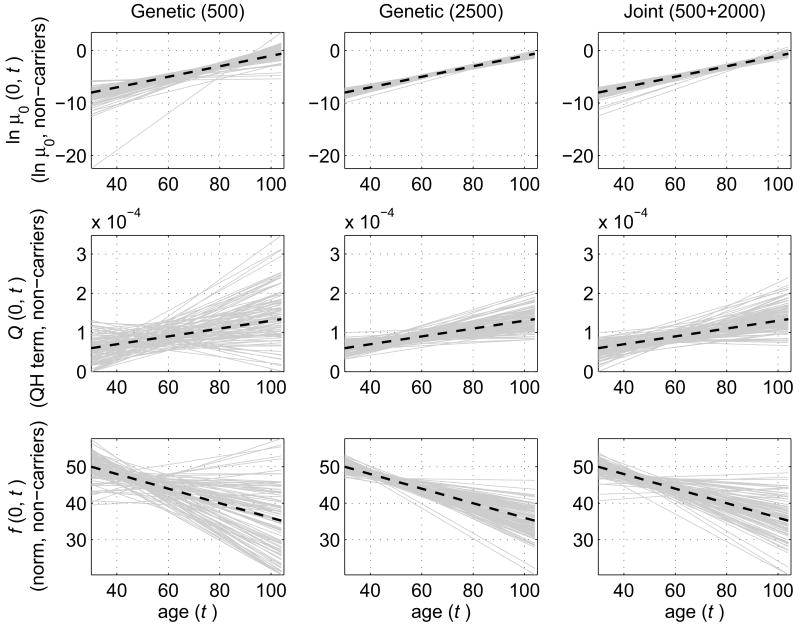
Fig. 2.
Simulation study: Comparison of estimates for non-carriers of a hypothetical allele (genotype) obtained in 100 simulated data sets by three methods. Left column: Estimates of age trajectories (solid grey lines) of logarithm of baseline hazard (ln μ0 (0,t) ), quadratic hazard (QH) terms (Q(0, t)), and age-dependent norms (f(0, t)) calculated using only sub-samples with genetic information (500 individuals). Middle column: Similar estimates when the entire sample (2500 individuals) contains genetic information. Right column: The estimates calculated using the joint analysis of genetic (500 individuals) and non-genetic (2000 individuals) data. Respective “true” trajectories used for simulation of data are shown as dashed black lines; t denotes age.
Table 1 and Figs. 1–2 show that the joint analysis of genetic and non-genetic data leads to a substantial increase in the accuracy of estimates. Standard deviations of parameter estimates are about two to four times larger in case of estimating a genetic sub-sample of 500 individuals compared to the joint analysis of genetic and non-genetic data. In case of estimating a genetic sub-sample of 500 individuals, some estimated trajectories of μ0 (Z,t), Q(Z, t) and f(Z, t), for both carriers (Fig. 1) and non-carriers (Fig. 2) substantially deviated from the “true” ones (those used for simulation of data). Thus, conclusions based on these estimates would be unreliable. However, the accuracy of estimates in the joint analysis (which is also based on genetic data for 500 individuals) is roughly comparable to that obtained in the analysis of genetic data on all 2500 individuals (i.e., maximizing the likelihood function (3) for the entire sample of 2500). Thus, a substantial increase in the accuracy of estimates can be achieved without the need of collecting additional genetic data on 2000 individuals.
Simulation studies S1–S3 in Supplementary Material provide more examples of applications of the approach in different situations.
4. Discussion
The entire research potential of available longitudinal data remains underused if only a genetic subset of data is involved in genetic analyses (ignoring the presence of non-genetic data). To be capable of using such a potential, the genetic model of longitudinal data must be extended to describe data in the non-genetic subgroup as well. Such an extension can be performed using methods of heterogeneity analyses (Vaupel and Yashin, 1985), taking into account that the non-genetic subgroup is a mixture of carriers of the same alleles or genotypes represented in the genetic group. The benefits of combining genetic and non-genetic data come from the presence of common parameters describing genetic and non-genetic subsets of these data. Our analyses show that the approach is capable of producing useful results in analyzing data on aging and mortality (Tan et al., 2002; Tan et al., 2001; Yashin et al., 1999; Yashin et al., 2000). The method is also useful in the analysis of data on longevity and incidence of diseases collected in longitudinal surveys (Yashin et al., 2007a). The approach assumed simple parametric models for genotype-specific hazard rates (such as Gompertz, linear, logistic or quadratic functions of age) and resulted in substantial improvement of the accuracy of statistical estimates (compared to the analysis of genetic data alone) without an increase in the size of the genetic sample.
The analyses performed in the papers discussed above do not allow for making conclusions about dynamic mechanisms generating estimated differences in genotype-specific hazards. Gaining knowledge about such mechanisms is a challenge to researchers in aging-related disciplines who still cannot come to conclusion concerning causes and regularities of aging-related deterioration in health/well-being/survival status in humans. The lack of consensus in interpretation of the results of experimental studies of aging resulted in the absence of a comprehensive theory and models describing systemic mechanisms generating longitudinal data on aging-related processes in humans. Traditionally, only subsets of such data, selected for studying a specific problem, are analyzed and the available mosaic details and findings on aging still did not form the entire picture of the regularities of aging-related changes in humans.
The model presented in this paper incorporates several promising concepts having a potential for describing and explaining substantial portions of aging-related changes in humans (age-specific physiological norms, allostasis and allostatic load, homeostenosis, decline in stress resistance with age, and stochasticity). Evidently, since all these variables characterize the same process of aging they should be mutually dependent. Nevertheless, they lack systemic practical applications to human data because typically not all such mechanisms (e.g., decline in stress resistance or allostatic load) are directly measured in longitudinal data available to date. Consequently, this hampers evaluation of the genetic contribution to the aging-related decline in the health/well-being status and the life span modulated by these mechanisms. The unification of these concepts in a comprehensive model of aging, health, and longevity is an important step towards the development of a systemic methodology in aging research. Incorporation of genetic information into this model permits evaluation of all these characteristics for carriers of different alleles (genotypes) to address new questions concerning genetic influence on the aging-related changes in humans, which were not possible to address before. For example, one can test the hypotheses about differences in age-trajectories of physiological norms for carriers and non-carriers of a specific allele/genotype (see examples in Simulation studies S1 in Supplementary Material). One can evaluate and verify pre-disease pathways by studying age patterns and components of allostatic load in carriers and non-carriers of allele/genotype (e.g., a larger difference between functions f1 and f for carriers would mean larger values of allostatic load in individuals carrying such allele/genotype). Genetic influence on age-related decline in adaptive capacity can be studied by comparing respective estimates of a(Z, t) for carriers and non-carriers (e.g., in a one-dimensional case, a faster decline of the absolute value of this function with age in carriers would indicate that the presence of such allele/genotype results in a faster decline in adaptive capacity with age). Comparison of the quadratic hazard terms (Q) for carriers and non-carriers allows for evaluation of genetic effect on stress resistance associated with deviation of selected physiological index from the age-specific norm (e.g., in a one-dimensional case, a faster increase of Q with age for carriers would indicate a faster decline in stress resistance in individuals with such allele/genotype). Testing hypotheses on differences in baseline hazards in carriers and non-carriers would help determine if there are any differences in mortality risks related to unobserved factors (i.e., those not involved in the quadratic term and represented by the respective stochastic process) in such individuals.
An important feature of the model is that it is capable of extracting genetic information from the entire sample of longitudinal data consisting of genetic and non-genetic sub-samples. This leads to a substantial increase in the accuracy of statistical estimates of genetic parameters compared to estimates based only on information from a genetic sub-sample and such an increase is achieved without collecting additional genetic data. Such models can be applied to analyses of any similar type of “incomplete” data, i.e., for any fixed (time-independent) discrete variable which is available only for a sub-sample of individuals from the entire data set.
The approach presented in this paper has some limitations. The model assumes that the genetic group is a random sample from the entire data set. However, in real longitudinal data this assumption may be violated. For example, only individuals with some specific characteristics (e.g., those without chronic diseases or disability or only those below some specific age) may be genotyped according to the design of the study. Similarly, those refusing to provide biological samples for analyses may have different health or disability status and health-related variables (resulting in different proportions of carriers of selected alleles or genotypes among non-responders) than those in the genetic sub-sample. The approaches to include probabilistic mechanisms generating such data into the model need to be elaborated. Further, longitudinal data usually consist of individuals from different cohorts. This may complicate analyses of morbidity and mortality in cases where the genetic structure of subsequent cohorts represented in the data is not similar (e.g., there is a substantial variation in frequencies of genotypes due to migration or other reasons). Joint analyses of genetic and non-genetic data that ignore the presence of such trends in the frequencies of genotypes may introduce bias in the results. Methods for evaluating such bias and approaches allowing for taking differences in genotypes’ frequencies into account in the analyses of genetic aspects of aging and longevity using mortality data are described by Yashin et al. (2007a).
Another limitation of the approach presented in the paper (as in any other parametric model) is that it assumes specific functional forms for equations describing the dynamics of physiological indices and the mortality/morbidity risk. Although the quadratic form of the hazard rates as a function of physiological indices is biologically justified by numerous empirical observations of J- or U-shapes of the risks, in real applications to longitudinal data the actual functional form of the risk function producing the observations is unknown. Also, other functions (such as physiological norms or adaptive capacity) cannot be empirically evaluated from the available data and their specific form is also unknown. Thus, a possible misspecification of the true mechanism generating the data can lead to biased estimates. Simulation study S3 in Supplementary Material provides an example of application of the method in case of a misspecified model with different structure of the equation describing the mortality risk. In this example, despite the different forms of the equation for the mortality risk in two models, the function f(Z, t) still has the meaning of the age-dependent norm for the physiological index represented by the process Yt. That is, it corresponds to the minimal mortality at respective ages in both the quadratic hazard and Cox models. In this case, the estimation procedures produced the estimates of respective parameters for the age-dependent norm f(Z, t) (i.e., estimates of the respective minimum of mortality) close to the actual values used for simulations despite the misspecification of the model. Additional studies are needed, however, to evaluate biases due to misspecification of models in other situations. Note also that the method presented here may be applied not only to the quadratic hazards as in equation (2). Other functional forms of the mortality rate as a function of physiological indices may be explored within the approach as well. For example, one can analyze various modifications of the proportional hazards model (see Simulation study S3 and Yashin et al., 2007c as examples of such models). Therefore, in applications of the method to longitudinal data it may be necessary to fit the data using models with different functional forms of the hazard rate and different functions in equation (1) and compare the models to define the best fitting one.
In the model presented in this paper, genetic information is included as a dichotomous variable (the presence/absence of an allele) for simplicity of presentation. In applications to genetic data, it may be necessary to explore different types of genetic models. For example, one may assume that aging-related characteristics represented in the equations (e.g., physiological norms, the decline in adaptive capacity, etc.) are different for different genotypes. Then, the functions in (1)–(2) are genotype-specific and respective random variable Z has 3 values (say, 0, 1, and 2 for genotypes aa, Aa, and AA). The extension of the estimation procedure to cover such situations is straightforward. However, it may result in smaller improvements in the power of statistical analyses due to an increased number of parameters and smaller sizes of (genotype-specific) genetic sub-samples. Similarly, extensions to analyses of multiple genes may run into the difficulties related to multiplicity of the parameters, which will reduce the reliability of estimates. Computational burden may be another limiting factor in analyses of high-dimensional models (those with a large number of indices represented by the process Yt) because the estimation procedure requires the solution of differential equations (6)–(7) at each step of maximization of the likelihood function.
The model presented in this paper assumes that all characteristics related to the process of aging depend only on age and genotype (e.g., those represented by functions f1(Z, t), f(Z, t), and a(Z, t)). Other covariates traditionally observed in longitudinal studies (such as socio-demographic and behavioral factors), as well as unobserved factors, may also influence the respective trajectories. The model can easily take this into account by explicitly incorporating observed factors into the parametric specification of respective functions, and estimating unknown parameters from the data. Another possible extension of the model is to describe the individual allostatic load and the age-dependent norms as unobserved randomly changing heterogeneity variables represented by stochastic differential equations.
Hidden heterogeneity in a population may substantially affect the shapes of population mortality rates (Vaupel and Yashin, 1985). Ignoring effects of such hidden heterogeneity (i.e., effects of some unobserved factors of genetic or non-genetic origin that affect mortality risks) can lead to erroneous conclusions concerning biological regularities of aging-related processes (Yashin et al., 2008b). Thus, an important generalization of the genetic SPM proposed in this paper would be to include the effects of hidden heterogeneity. A version of the SPM that takes the presence of hidden heterogeneity into account was elaborated recently (Yashin et al., 2008b).
Another direction is to develop and investigate a model describing the joint age-dynamics of a physiological state (modeled by a continuous process) and health/well-being status (represented by a discrete variable) in humans. Such a description would allow one to analyze data that include systems of longitudinal measurements performed under different observational plans on the same individuals. The introduction of a finite state (discrete) component for health/well-being state permits investigation of the dynamics of physiological and other indices considered before and after major health or disability events. It will allow for evaluating the role of physiological trajectories in the age-related increase in the risks of developing a disease, disability and death and uncovering pre-disease physiological pathways and differences in these characteristics among carriers of different genotypes.
Recently, data on genome-wide association studies (GWAS) are becoming available for participants of large-scale longitudinal surveys, for example the Framingham Heart Study participants from all three generations (over 9,300 individuals) have been genotyped in a 550,000 SNPs GWAS (the FHS SHARe project: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v4.p2). Such data sets contain all information that is needed for application of the approach suggested in this paper: genetic information (SNPs) available for a part of the sample and a numerous physiological variables measured longitudinally for individuals with available genetic information and for the rest of the sample, i.e., those who were not genotyped in the GWAS, and the history of morbidity events for all individuals (as the genotyping has been performed quite recently, there may be few or no mortality events in the genetic sub-sample). Although applications of the presented approach to routine analyses of entire GWAS data may be infeasible due to excessive computational burden (let alone the usual multiple comparison problem of GWAS), it may still prove to be useful for analyses of candidate SNPs and their connection to various health- and aging-related phenotypes. For example, major genetic pathways that regulate proliferation, apoptosis, replicative senescence, and autophagy, as well as genes that govern the interactions among these pathways (that is, SNPs located within/near the genes belonging to the above pathways) may be plausible candidates for analyses of their associations with various phenotypes of (healthy) aging within a framework of the presented model.
Supplementary Material
Acknowledgments
This work was supported by grants R01AG030612, R01AG027019, R01AG028259, and 5P01AG008761 from the National Institute on Aging. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison DB, Faith MS, Heo M, Kotler DP. Hypothesis concerning the U-shaped relation between body mass index and mortality. Am J Epidemiol. 1997;146:339–349. doi: 10.1093/oxfordjournals.aje.a009275. [DOI] [PubMed] [Google Scholar]
- Boutitie F, Gueyffier F, Pocock S, Fagard R, Boissel JP. J-shaped relationship between blood pressure and mortality in hypertensive patients: New insights from a meta-analysis of individual-patient data. Ann Intern Med. 2002;136:438–448. doi: 10.7326/0003-4819-136-6-200203190-00007. [DOI] [PubMed] [Google Scholar]
- Breslow NE, Cain KC. Logistic regression for two-stage case-control data. Biometrika. 1988;75:11–20. [Google Scholar]
- Breslow NE, Holubkov R. Weighted likelihood, pseudo-likelihood and maximum likelihood methods for logistic regression analysis of two-stage data. Stat Med. 1997a;16:103–116. doi: 10.1002/(sici)1097-0258(19970115)16:1<103::aid-sim474>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Breslow NE, Holubkov R. Maximum likelihood estimation of logistic regression parameters under two-phase, outcome-dependent sampling. J Royal Stat Soc B. 1997b;59:447–461. [Google Scholar]
- Bureau A, Diallo MS, Ordovas JM, Cupples LA. Estimating interaction between genetic and environmental risk factors: Efficiency of sampling designs within a cohort. Epidemiology. 2008;19:83–93. doi: 10.1097/EDE.0b013e31815c4d0e. [DOI] [PubMed] [Google Scholar]
- Cain KC, Breslow NE. Logistic regression analysis and efficient design for two–stage studies. Am J Epidemiol. 1988;128:1198–1206. doi: 10.1093/oxfordjournals.aje.a115074. [DOI] [PubMed] [Google Scholar]
- Chatterjee N, Chen YH. Maximum likelihood inference on a mixed conditionally and marginally specified regression model for genetic epidemiologic studies with two-phase sampling. J Royal Stat Soc B. 2007;69:123–142. [Google Scholar]
- Dawber TR, Meadors GF, Moore FE. Epidemiological approaches to heart disease: The Framingham Study. Am J Public Health. 1951;41:279–286. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberger AL, Peng CK, Lipsitz LA. What is physiologic complexity and how does it change with aging and disease? Neurobiol Aging. 2002;23:23–26. doi: 10.1016/s0197-4580(01)00266-4. [DOI] [PubMed] [Google Scholar]
- Hall DM, Xu L, Drake VJ, Oberley LW, Oberley TD, Moseley PL, Kregel KC. Aging reduces adaptive capacity and stress protein expression in the liver after heat stress. J Appl Physiol. 2000;89:749–759. doi: 10.1152/jappl.2000.89.2.749. [DOI] [PubMed] [Google Scholar]
- Karlamangla AS, Singer BH, Seeman TE. Reduction in allostatic load in older adults is associated with lower all-cause mortality risk: MacArthur studies of successful aging. Psychosom Med. 2006;68:500–507. doi: 10.1097/01.psy.0000221270.93985.82. [DOI] [PubMed] [Google Scholar]
- Kulminski AM, Arbeev KG, Kulminskaya IV, Ukraintseva SV, Land K, Akushevich I, Yashin AI. Body mass index and nine-year mortality in disabled and nondisabled older U.S. Individuals. J Am Geriatr Soc. 2008;56:105–110. doi: 10.1111/j.1532-5415.2007.01494.x. [DOI] [PubMed] [Google Scholar]
- Kuzuya M, Enoki H, Iwata M, Hasegawa J, Hirakawa Y. J-shaped relationship between resting pulse rate and all-cause mortality in community-dwelling older people with disabilities. J Am Geriatr Soc. 2008;56:367–368. doi: 10.1111/j.1532-5415.2007.01512.x. [DOI] [PubMed] [Google Scholar]
- Lewington S, Clarke R, Qizilbash N, Peto R, Collins R. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C-elegans. Curr Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- Manton KG, Yashin AI. Odense Monograph on Population Aging No. 7. Odense University Press; Odense, Denmark: 2000. Mechanisms of Aging and Mortality: A Search for New Paradigms. [Google Scholar]
- MathWorks Inc. Optimization Toolbox™ 4 User’s Guide. The MathWorks, Inc.; Natick, MA: 2008. [Google Scholar]
- Mazza A, Zamboni S, Rizzato E, Pessina AC, Tikhonoff V, Schiavon L, Casiglia E. Serum uric acid shows a J-shaped trend with coronary mortality in non-insulin-dependent diabetic elderly people. The CArdiovascular STudy in the ELderly (CASTEL) Acta Diabetol. 2007;44:99–105. doi: 10.1007/s00592-007-0249-3. [DOI] [PubMed] [Google Scholar]
- McEwen BS, Wingfield JC. The concept of allostasis in biology and biomedicine. Horm Behav. 2003;43:2–15. doi: 10.1016/s0018-506x(02)00024-7. [DOI] [PubMed] [Google Scholar]
- Okumiya K, Matsubayashi K, Wada T, Fujisawa M, Osaki Y, Doi Y, Yasuda N, Ozawa T. A U-shaped association between home systolic blood pressure and four-year mortality in community-dwelling older men. J Am Geriatr Soc. 1999;47:1415–1421. doi: 10.1111/j.1532-5415.1999.tb01559.x. [DOI] [PubMed] [Google Scholar]
- Palatini P. Need for a revision of the normal limits of resting heart rate. Hypertension. 1999;33:622–625. doi: 10.1161/01.hyp.33.2.622. [DOI] [PubMed] [Google Scholar]
- Protogerou AD, Safar ME, Iaria P, Safar H, Le Dudal K, Filipovsky J, Henry O, Ducimetiere P, Blacher J. Diastolic blood pressure and mortality in the elderly with cardiovascular disease. Hypertension. 2007;50:172–180. doi: 10.1161/HYPERTENSIONAHA.107.089797. [DOI] [PubMed] [Google Scholar]
- Scott AJ, Wild CJ. Maximum likelihood for generalised case-control studies. Journal of Statistical Planning and Inference. 2001;96:3–27. [Google Scholar]
- Scott AJ, Lee AJ, Wild CJ. On the Breslow-Holubkov estimator. Lifetime Data Anal. 2007;13:545–563. doi: 10.1007/s10985-007-9054-0. [DOI] [PubMed] [Google Scholar]
- Seeman TE, McEwen BS, Rowe JW, Singer BH. Allostatic load as a marker of cumulative biological risk: MacArthur studies of successful aging. Proc Natl Acad Sci U S A. 2001;98:4770–4775. doi: 10.1073/pnas.081072698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan QH, De Benedictis G, Ukraintseva SV, Franceschi C, Vaupel JW, Yashin AI. A centenarian-only approach for assessing gene-gene interaction in human longevity. Europ J Hum Genet. 2002;10:119–124. doi: 10.1038/sj.ejhg.5200770. [DOI] [PubMed] [Google Scholar]
- Tan QH, Yashin AI, Bladbjerg EM, de Maat MPM, Andersen-Ranberg K, Jeune B, Christensen K, Vaupel JW. Variations of cardiovascular disease associated genes exhibit sex-dependent influence on human longevity. Exp Gerontol. 2001;36:1303–1315. doi: 10.1016/s0531-5565(01)00102-4. [DOI] [PubMed] [Google Scholar]
- Troiano RP, Frongillo EA, Sobal J, Levitsky DA. The relationship between body weight and mortality: A quantitative analysis of combined information from existing studies. Int J Obesity. 1996;20:63–75. [PubMed] [Google Scholar]
- Troncale JA. The aging process: Physiologic changes and pharmacologic implications. Postgrad Med. 1996;99:111–114. 120–122. [PubMed] [Google Scholar]
- Ukraintseva SV, Yashin AI. Individual aging and cancer risk: How are they related? Demographic Research. 2003;9:163–196. [Google Scholar]
- Vaupel JW, Yashin AI. Heterogeneity’s ruses: some surprising effects of selection on population dynamics. Amer Statistician. 1985;39:176–185. [PubMed] [Google Scholar]
- Westin S, Heath I. Thresholds for normal blood pressure and serum cholesterol. Br Med J. 2005;330:1461–1462. doi: 10.1136/bmj.330.7506.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witteman JCM, Grobbee DE, Valkenburg HA, Vanhemert AM, Stijnen T, Burger H, Hofman A. J-shaped relation between change in diastolic blood-pressure and progression of aortic atherosclerosis. Lancet. 1994;343:504–507. doi: 10.1016/s0140-6736(94)91459-1. [DOI] [PubMed] [Google Scholar]
- Woodbury MA, Manton KG. Random-walk model of human mortality and aging. Theor Popul Biol. 1977;11:37–48. doi: 10.1016/0040-5809(77)90005-3. [DOI] [PubMed] [Google Scholar]
- Yashin AI. Dynamics in survival analysis: Conditional Gaussian property vs. Cameron-Martin formula. In: Krylov NV, et al., editors. Statistics and Control of Stochastic Processes. Springer; New York: 1985. pp. 446–475. [Google Scholar]
- Yashin AI, Manton KG. Effects of unobserved and partially observed covariate processes on system failure: A review of models and estimation strategies. Statistical Science. 1997;12:20–34. [Google Scholar]
- Yashin AI, Arbeev KG, Ukraintseva SV. The accuracy of statistical estimates in genetic studies of aging can be significantly improved. Biogerontology. 2007a;8:243–255. doi: 10.1007/s10522-006-9072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashin AI, Akushevich IV, Arbeev KG, Akushevich L, Ukraintseva SV, Kulminski A. Insights on aging and exceptional longevity from longitudinal data: novel findings from the Framingham Heart Study. Age. 2006;28:363–374. doi: 10.1007/s11357-006-9023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashin AI, Arbeev KG, Akushevich I, Kulminski A, Akushevich L, Ukraintseva SV. Stochastic model for analysis of longitudinal data on aging and mortality. Math Biosci. 2007b;208:538–551. doi: 10.1016/j.mbs.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashin AI, Arbeev KG, Kulminski A, Akushevich I, Akushevich L, Ukraintseva SV. Cumulative index of elderly disorders and its dynamic contribution to mortality and longevity. Rejuvenation Research. 2007c;10:75–86. doi: 10.1089/rej.2006.0500. [DOI] [PubMed] [Google Scholar]
- Yashin AI, Arbeev KG, Kulminski A, Akushevich I, Akushevich L, Ukraintseva SV. Health decline, aging and mortality: how are they related? Biogerontology. 2007d;8:291–302. doi: 10.1007/s10522-006-9073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashin AI, Arbeev KG, Kulminski A, Akushevich I, Akushevich L, Ukraintseva SV. What age trajectories of cumulative deficits and medical costs tell us about individual aging and mortality risk: Findings from the NLTCS-Medicare data. Mech Ageing Dev. 2008a;129:191–200. doi: 10.1016/j.mad.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashin AI, Arbeev KG, Akushevich I, Kulminski A, Akushevich L, Ukraintseva SV. Model of hidden heterogeneity in longitudinal data. Theor Popul Biol. 2008b;73:1–10. doi: 10.1016/j.tpb.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashin AI, De Benedictis G, Vaupel JW, Tan Q, Andreev KF, Iachine IA, Bonafe M, DeLuca M, Valensin S, Carotenuto L, Franceschi C. Genes, demography, and life span: The contribution of demographic data in genetic studies on aging and longevity. Am J Hum Genet. 1999;65:1178–1193. doi: 10.1086/302572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashin AI, De Benedictis G, Vaupel JW, Tan Q, Andreev KF, Iachine IA, Bonafe M, Valensin S, De Luca M, Carotenuto L, Franceschi C. Genes and longevity: Lessons from studies of centenarians. J Gerontol A Biol Sci Med Sci. 2000;55:B319–B328. doi: 10.1093/gerona/55.7.b319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.