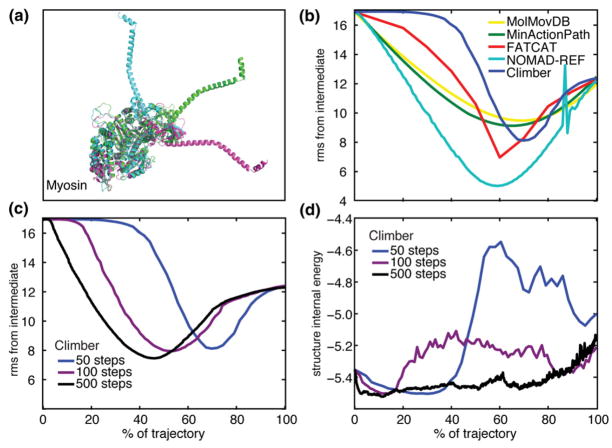
Fig. 1.
(a) The three crystallographic structures of scallop myosin II. (b) The Cα rmsd (in angstroms) of the interpolated myosin structures from the crystallographic intermediate structure is shown for MolMovDB (yellow), MinActionPath (green), FATCAT (red), NOMAD-Ref (cyan), and Climber with Ncycle = 50 (blue). For clarity, the controls are not shown because they never come closer to the intermediate compared to the endpoints. (c) The Cα rmsd from the crystallographic intermediate using Ncycle =50 (blue), 100 (violet), and 500 (black) as input to the Climber morphing method. (d) The internal energy of the intermediate structures using Ncycle =50, 100, and 500 in the Climber method. A longer interpolation with a smaller desired cRMS change per step allows Climber to find the low-energy pathway. An increase in the energy at the end of the morph is due to the difficulty of repacking the side chains into the compact final structure.