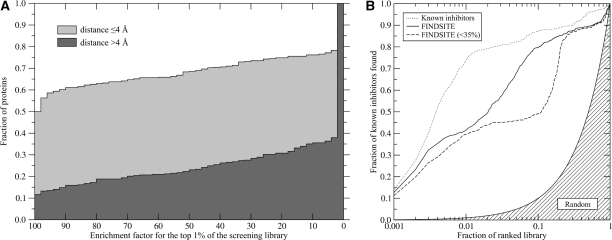
Figure 4:
(A) Using FINDSITE selected ligand templates, cumulative distribution of enrichment factors from the ligand-based virtual screening experiment against the KEGG compound library. Depending on the whether the distance between the top-ranked pocket and the center of mass of the native ligand ≤ 4 Å and >4 Å, target proteins are divided into two subsets. (B) Enrichment behavior in virtual screening for HIV-1 protease inhibitors using ligands predicted by FINDSITE from either homologous or weakly homologous threading templates compared to that using known inhibitors and random ligand ranking.