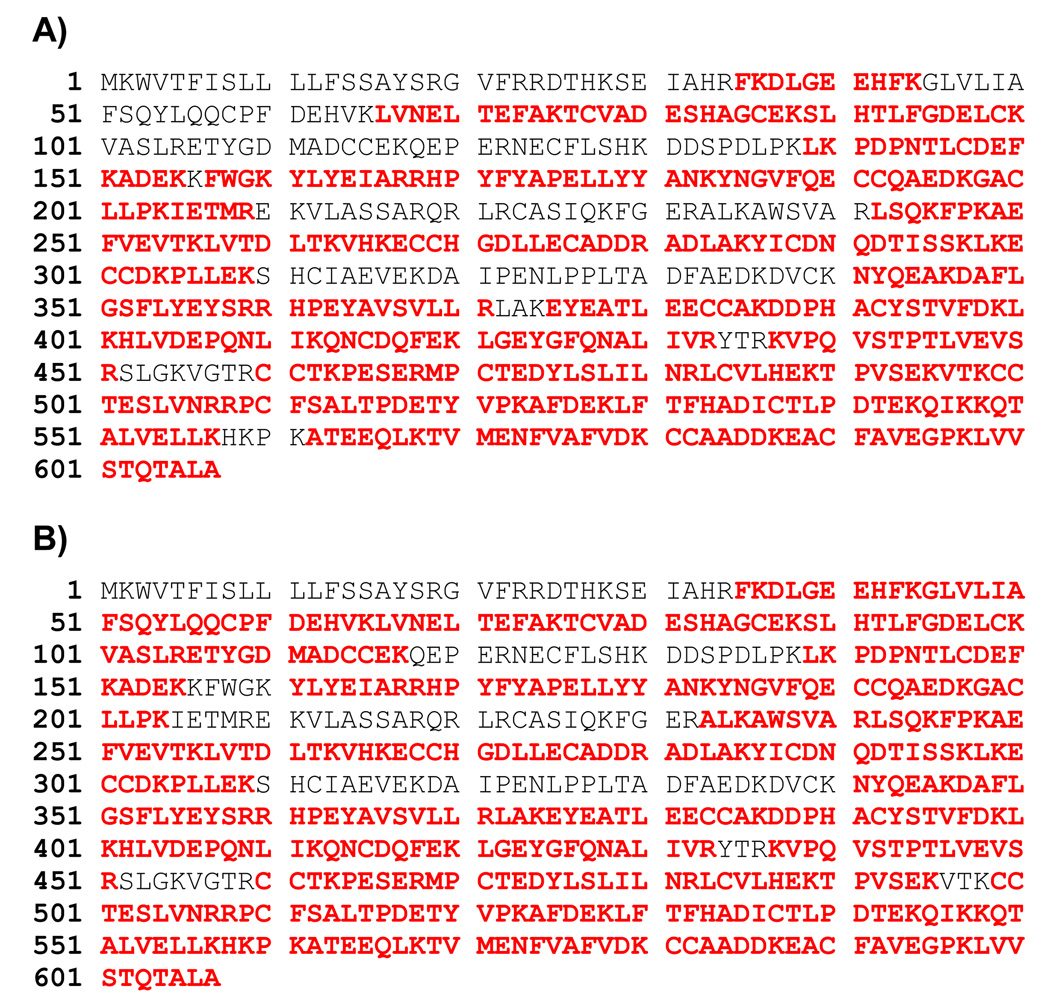
Figure 6.
Amino acid sequence coverage of BSA by in-solution digestion of BSA by rSET (A) and modified porcine trypsin (B). LC-MS/MS data of 5 h BSA digests shown in Figure 5 were subjected to Mascot search against a database containing solely BSA amino acid sequence. Mascot database search software version 2.1.04 (Matrix Science, London, UK) was used. Mass tolerances for protein identification for precursor ion was set to 20 ppm and for product ion set to 1 Da. Strict trypsin specificity was applied, allowing for one missed cleavage. Only peptides with a minimum score of 20 were considered as significant. Identified peptides were shown in bold red.