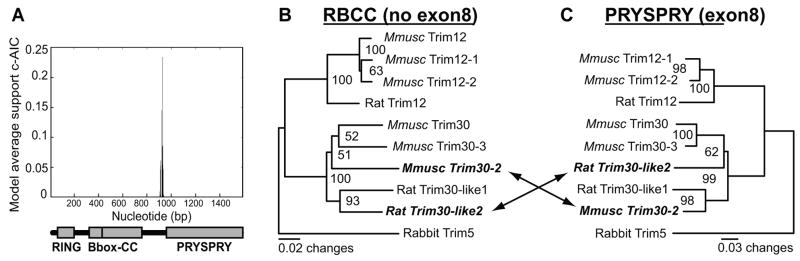
Figure 3. Recombination among rodent Trim5 genes.
(A) The online GARD tool identifies a breakpoint upstream of the PRYSPRY (exon8) domain for the full-length canonical Trim5α sequences from mice, rats and rabbits. The second-order Akaike Information Criterion (c-AIC) value is shown on the y axis as a measure to show the goodness of fit of this location of the breakpoint compared to other locations identified by GARD (not shown). The model average support value (numbers on the y axis) by itself has no meaning; they are used by GARD to identify the best single breakpoint among a series of possible breakpoints. Nucleotide position is indicated on the x axis. DNA sequences of the (B) RBCC domains (excluding exon8), and of the (C) PRYSPRY (exon8) domain for rodent Trim5 homologs were used to construct a maximum likelihood tree. Rabbit Trim5α was used as an outgroup. Sequences that showed evidence of recombination are shown in bold text. Mmusc stands for Mus musculus. Mmusc Trim12 was excluded because it lacks a PRYSPRY domain (exon8). Scale for branch length is shown below the tree.