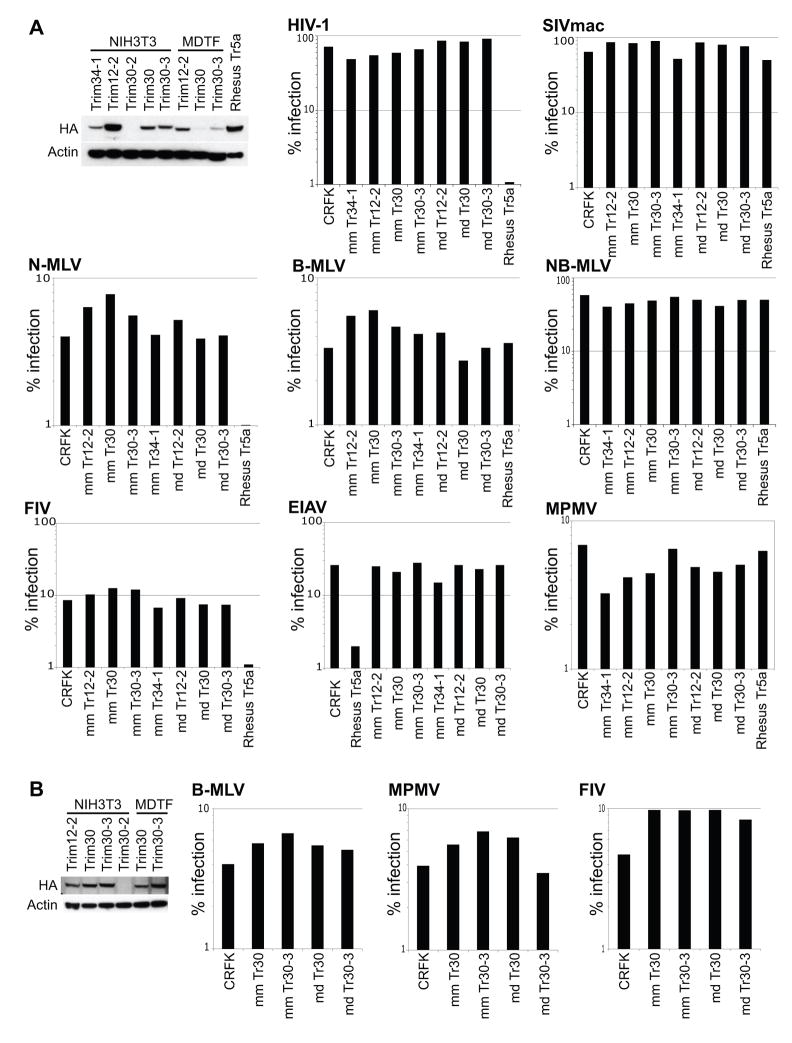
Figure 6. Retroviruses used in our assays were not restricted by mouse Trim5 homologs.
CRFK cell lines were transduced to stably express Trim5 homologs containing all four of the canonical domains of Trim5α, cloned from NIH3T3 and MDTF cells, representing Mus musculus and Mus dunni respectively. Stable cell lines for Trim34-1 from NIH3T3 cells and Rhesus Trim5α were also generated. Each Trim gene was cloned with an N-terminal HA tag for detection via immunoblotting. (A and B) 50 ug of lysates per well, immunoblotted with anti-HA to check for expression of each Trim5 homolog. Anti-actin was used as a loading control. A second attempt at stable cell line generation was needed to boost the levels of MDTF Trim30 and Trim30-3 (B). We were unable to generate CRFK cells stably expressing Trim30-2 from either NIH3T3 or MDTF cells.
(A and B) Each stable cell line expressing one of the listed Trim genes was challenged with the list of retroviruses at sufficient amounts to give non-saturating levels of infection. Each virus is generated to express GFP upon infection. These retroviruses were used because they represent a broad range of viruses restricted by antiviral Trim5 orthologs from primates, cows and rabbits. HIV-1 (human immunodeficiency virus-1), SIVmac (simian immunodeficiency virus macaque strain), MPMV (mason-pfizer monkey virus), N-MLV (N-tropic murine leukemia virus), B-MLV (B-tropic murine leukemia virus), NB-MLV (N- and B-tropic murine leukemia virus), FIV (feline immunodeficiency virus), EIAV (equine infectious anemia virus). Percentage of cells that are infected are plotted on the y-axis as a log-scale.