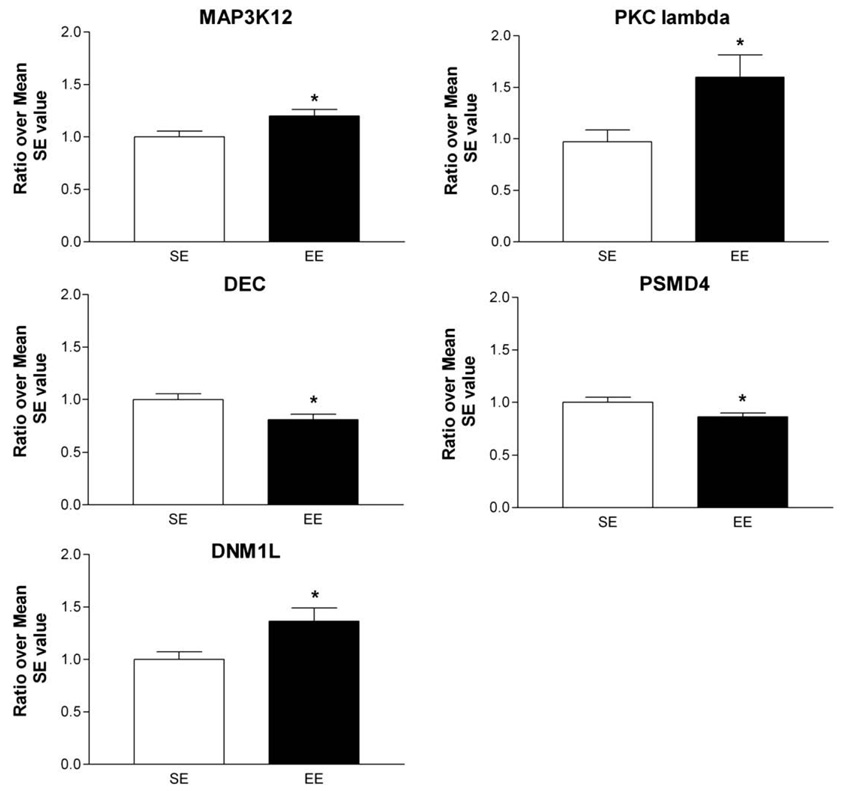
Fig. 2.
Quantitative RT-PCR analysis of some transcripts affected by EE. The expression of genes, selected from various families of genes for their potential involvement in EE's effects on striatal plasticity, was confirmed by RT-PCR. Selected genes were: Mitogen-activated protein kinase kinase kinase 12 (MAP3K12), Protein kinase C lambda (PKC lambda), Decorin (DEC), Proteasome 26S subunit non-ATPase 4 (PSMD4), and Dynamin 1-like (DNM1L). For all genes examined, statistically significant changes were observed in the same direction as arrays results. The data were obtained from 8 mice per group and determined individually. The amount of each product was normalized by GAPDH value and then a ratio over mean SE value was obtained. Values represent means±SEM. Student t-test, *p< 0.05 different from SE control.