Abstract
Phospholipid transfer protein (PLTP), one of the key lipid transfer proteins in plasma and cerebrospinal fluid, is nearly ubiquitously expressed in cells and tissues. Functions of secreted PLTP have been extensively studied. However, very little is known about potential intracellular PLTP functions. In the current study, we provide evidence for PLTP localization in the nucleus of cells that constitutively express PLTP (human neuroblastoma cells, SK-N-SH; and human cortical neurons, HCN2) and in cells transfected with human PLTP (Chinese hamster ovary and baby hamster kidney cells). Furthermore, we have shown that incubation of these cells with leptomycin B (LMB), a specific inhibitor of nuclear export mediated by chromosome region maintenance 1 (CRM1), leads to intranuclear accumulation of PLTP, suggesting that PLTP nuclear export is CRM1-dependent. We also provide evidence for entry of secreted PLTP into the cell and its translocation to the nucleus, and show that intranuclear PLTP is active in phospholipid transfer. These findings suggest that PLTP is involved in novel intracellular functions.
Keywords: Phospholipid transfer protein, PLTP, nuclear localization, neuronal cells, leptomycin B
Introduction
Phospholipid transfer protein (PLTP) is a versatile transfer protein with predicted protein molecular mass of approximately 55 kDa with signal peptide, or 53 kDa without [1]. Posttranslational modifications, such as N-glycosylation, account for the difference observed between the predicted molecular mass, 53 kDa, and observed mass of 81 kDa of the secreted protein [1]. Secreted PLTP is bound to lipoprotein particles, and associates with apolipoprotein A-I (apoA-I) and E (apoE), and with lipoproteins containing other apolipoproteins [2, 3]. PLTP functions have been associated with the metabolism of lipoproteins, particularly high-density lipoproteins (HDL) (reviewed in [4]). PLTP has been shown to modulate phospholipid and cholesterol transfer between lipoproteins, as well as between lipoproteins and cells [5–7]. PLTP also transfers apolipoprotein E (apoE), α-tocopherol and other molecules such as cerebrosides and diacylglycerides [7–11].
PLTP is expressed by nearly all types of cells and tissues, with wide variation in expression patterns ([5, 12] and unpublished data). For example, neighboring neurons in Cornu Amonis (CA) layers 1 and 2 in human hippocampus differ significantly in PLTP expression [12]. Interestingly, neurons in the CA1 layer, which express significantly lower levels of PLTP than other hippocampal neurons, are particularly sensitive to oxidative stress [12, 13], and PLTP is one of the key molecules for maintenance of the brain anti-oxidative properties through regulation of tissue levels of α-tocopherol [11]. These findings, along with nearly ubiquitous, yet differential expression of PLTP in other cells and tissues suggest that PLTP has physiological roles other than purely extracellular functions related to lipoprotein metabolism. Although the role of PLTP as a secretory protein has been extensively studied, nothing is known about the possible intracellular functions of this protein.
In our immunohistochemical studies of the human brain [12], we noticed intranuclear staining in neuronal cells using several different antibodies against PLTP (unpublished data). We tested these initial observations in order to establish whether PLTP is indeed present in the nucleus. In this study we provide evidence for PLTP presence and activity in the nuclear compartment and discuss motifs that could be involved in PLTP nuclear import and export, implying that PLTP has novel intracellular functions beyond that of extracellular lipid transport.
Material and Methods
Materials
Dulbecco modified cell culture media (DMEM) were purchased from Lonza (Walkersville, MD). Chinese hamster ovary cells prepared for Flp-In vectors, Flp-In vectors and specialized growth media were purchased from Invitrogen (Carlsbad, CA). Fetal calf serum was obtained from HyClone (Logan, UT). Mutagenesis kits were purchased from Qiagen (Valencia, CA). Leptomycin B (LMB), bovine skin collagen, Triton X-100 and antibodies against lamin A were purchased from Sigma-Aldrich (St. Louis, MO). Nunc Lab-Tech chamber slides and other cell culture plastic-ware were purchased from VWR (West Chester, PA). Methanol-free formaldehyde (16%) was purchased from Polysciences (Warrington, PA). Monoclonal antibody against PLTP was produced at the Northwest Lipid Research Laboratory, in collaboration with Dr. Marcovina [12]. Secondary antibodies for Western blotting were purchased from eBioscience (San Diego, CA). Antibodies against TATA binding protein (TBP) for Western loading control were purchased from Abcam (Cambridge, MA). Biotinylated secondary antibodies (horse anti-mouse), Vectastain ELITE ABC, NovaRED and DAB kits, horse serum, hematoxylin, AquaMount and VectaShield mounting media were purchased from Vector Laboratories (Burlingame, CA). Fluorescently labeled secondary antibodies, chicken anti-mouse AlexaFluor 488 and goat anti-rabbit AlexaFluor 594 antibodies, and Image-iT FX signal enhancer were from Molecular Probes/Invitrogen (Eugene, OR). PLTP tagged with enhanced green fluorescent protein (PLTP-EGFP) and mock vectors were a kind gift from Dr. Vesa Olkkonen, National Public Health Institute, Helsinki, Finland [14]. Cytoplasmic and nuclear proteins were isolated using NE-PER kit with addition of protease inhibitor cocktail, Halt (Pierce, Rockford, IL).
Cell culture
Experiments were conducted in human neuroblastoma cells (SK-N-SH), human cortical neurons (HCN2; ATCC CRL-10743), Chinese hamster ovary (CHO) and baby hamster kidney (BHK) cells. Cells were incubated in 5% CO2, at 37°C, supplemented with 5% (SK-N-SH), 10% (CHO and BHK) or 15% (HCN2) serum in growth media. For standard immunocytochemistry and fluorescent live-cell imaging cells were plated in 8-chamber slides, while cells prepared for confocal microscopy were grown on collagen-treated coverslips in six-well plates. Cells were incubated without or with 10 ng/ml of LMB for up to 24 hours. All experiments were performed in serum-free media, thus eliminating potential external source of PLTP, conducted in triplicate, and replicated multiple times.
PLTP mutagenesis
Site-directed mutagenesis was conducted using wild-type (WT) PLTP cDNA (Day et al. 1994) as a template, using QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Site-directed in vitro mutagenesis was accomplished by hybridizing a plasmid containing the PLTP insert (human PLTP C-his tag/pcDNA5/FRT) to oligonucleotides which are complementary to the target template except for a region of mismatch near the center, containing the desired serine-to-alanine mutations. We mutated the following residues in PLTP: S49A, S79A, S102A, and S128A within the putative N-glycosylation motifs (N/X/S-T; numbering based on mature protein without signal peptide). All created cDNA were verified by digestion and sequencing prior to transfection.
PLTP expression
Recombinant PLTP (rPLTP) constructs containing His tag were transfected into CHO Flp-In-Ready cells (Invitrogen) using Lipofectamine 2000, and clones were selected using Hygromycin B (0.6 mg/ml; Invitrogen). PLTP-EGFP constructs were transfected into BHK cells using Lipofectamine 2000, and positive clones were selected using G418 (1 mg/ml; Invitrogen). Highest expressing clones for PLTP-EGFP were selected by measurement of fluorescence (excitation/emission 488/535 nm), and all PLTP clones (wild-type His-tagged and PLTP-EGFP, as well as mutant PLTP) were confirmed by Western blot analysis using PLTP-specific monoclonal antibodies, and by measurement of PLTP phospholipid transfer activity in an in vitro assay [15].
Immunocytochemistry
Following incubation, cells prepared for standard, chromogenic ICC were fixed with 95% ethanol for 10 minutes, washed and incubated for 30 minutes with 3% hydrogen peroxide to reduce endogenous peroxidase. Slides were blocked with horse serum for 20 minutes, and incubated with mouse anti-PLTP antibody (20 μg/ml) overnight at 4°C, followed by biotinylated horse anti-mouse antibody (5 μg/ml) for thirty minutes. The signal was amplified using Vectastain ELITE ABC reagents for 30 minutes, and stained using DAB or NovaRED kits. The slides were counterstained with hematoxylin, coverslipped and observed under Olympus microscope. Digital images were created using Nikon E990 digital camera. Cells prepared for confocal microscopy were grown on coverslips, incubated without or with 10 ng/ml LMB for up to 24 hours, fixed using 3.7% formaldehyde for 15 minutes, and incubated with primary antibodies against PLTP (mouse; 20 μg/ml) and Lamin A (rabbit; 2.5 μg/ml), as a nuclear marker, at 4°C overnight. Alexa Fluor secondary antibodies (chicken anti-mouse Alexa Fluor 488, 5 μg/ml; and goat anti-rabbit Alexa Fluor 594, 3 μg/ml) for 45 minutes, rinsed with PBS and mounted using VectaShield HardSet mounting medium. The slides were observed under confocal Zeiss LSM 510 Meta microscope, using water-immersion objective, and recorded using Zeiss LSM 510 software, version 4.1 SP1.
Live cell imaging Live cells transfected with PLTP-EGFP and grown in 8-chamber slides in phenol red-free media, incubated without or with LMB (10 ng/ml), were observed under Zeiss Axiovert inverted fluorescent microscope, using QColor 3 digital camera and QCapture Pro software.
PLTP re-entry experiment: Phenol red-free media from control BHK cells and cells transfected with PLTP-EGFP were collected after 24-hour incubation, centrifuged to remove cells, concentrated using 30 kDa cut-off Amicon Ultra-4 centrifugal filter device, and added to control
BHK cells incubated with vehicle (70% methanol) or 10 ng/ml LMB for 24 hours. Following incubation, cells were fixed with formaldehyde and observed under confocal microscope.
Western blot
Plated cells were incubated in the presence or absence of LMB from 1 to 24 hours. Following incubation, cells were washed, scraped and cytoplasmic and nuclear extract proteins were isolated using NE-PER kit (Pierce) following the manufacturer’s instructions. Briefly, cells were lysed using CERI buffer containing protease inhibitor cocktail (Halt protease inhibitor cocktail, Pierce). Following cell lysis, CERII buffer was added, and cells were incubated, vortexed and centrifuged at 16,000 × g at 4°C. Cytoplasmic extract was separated and stored at −80°C until use. Nuclear pellet was resuspended in NER buffer containing protease inhibitor cocktail. Lysed nuclei were subjected to centrifugation, and supernatant containing nuclear extract was stored at −80°C until use. Equal amounts of nuclear proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 4–12% Criterion gel (Bio-Rad, Hercules, CA), transferred to nitrocellulose membrane using semi-dry transfer system (BioRad), and subjected to Western blot analysis using monoclonal anti-PLTP antibody (1 μg/ml, overnight incubation at 4°C), and anti-mouse secondary antibody that reacts exclusively with non-denatured immunoglobulin, thus eliminating false positive results (TrueBlot, eBioscience, San Diego, CA; 0.2 μg/ml, 2 hour incubation at room temperature). Loading control was assessed using anti-TBP (Abcam; 0.08 μg/ml). Positive bands were visualized using West Femto SuperSignal kit (Pierce). Membranes were digitally scanned using Kodak CF400 Gel Imager.
Phospholipid transfer activity assay
PLTP-mediated phospholipid transfer activity was assessed using PLTP phospholipid transfer activity assay [15]. Nuclear proteins were assayed for the ability to transfer C14-labeled phospholipid from donor liposome particles to the acceptor particles (plasma HDL without measurable PLTP activity). All values were adjusted for the background transfer, and PLTP activity was calculated as percent of phospholipid transfer per nanogram of total nuclear protein per hour (%/ng/h). PLTP-mediated phospholipid transfer activity was confirmed using anti-PLTP antibody inhibition. Nuclear extracts were incubated overnight at 4°C with 1 μg of chicken anti-PLTP antibody [16], using the minimal concentration that elicited complete inhibition of PLTP activity in the control plasma sample. Samples were then centrifuged for twenty minutes at low speed at 4°C. The supernatant was evaluated for phospholipid transfer activity. Control chicken IgG was used as negative control, and assay performance was evaluated using a plasma sample with known PLTP activity inhibition under identical conditions.
Computer analyses
Putative nuclear export motifs in the PLTP sequence were evaluated using online Nuclear Export Server (NetNES 1.1; http://www.cbs.dtu.dk/services/NetNES/), and nuclear import motifs were assessed using an online tool (http://cubic.bioc.columbia.edu/cgi/var/nair/resonline.pl). Following these analyses, PLTP protein sequence was further evaluated for nuclear export and nuclear import sequences based on the published consensus sequences [17–21].
Statistical analyses
Statistical analyses were performed using Statistica for Windows, StatSoft, Inc., 2000 (Tulsa, OK). P-values <0.05 were considered statistically significant.
Results
To establish whether or not PLTP is present in the nuclei of neurons, we isolated nuclear proteins from SK-N-SH neuroblastoma cells, and analyzed them by Western blot. The analysis suggested that PLTP is indeed present in the nuclei (Figure 1a). Similar results were obtained using a nuclear extract isolated from human cortical neurons (HCN2; Figure 1b).
Figure 1.
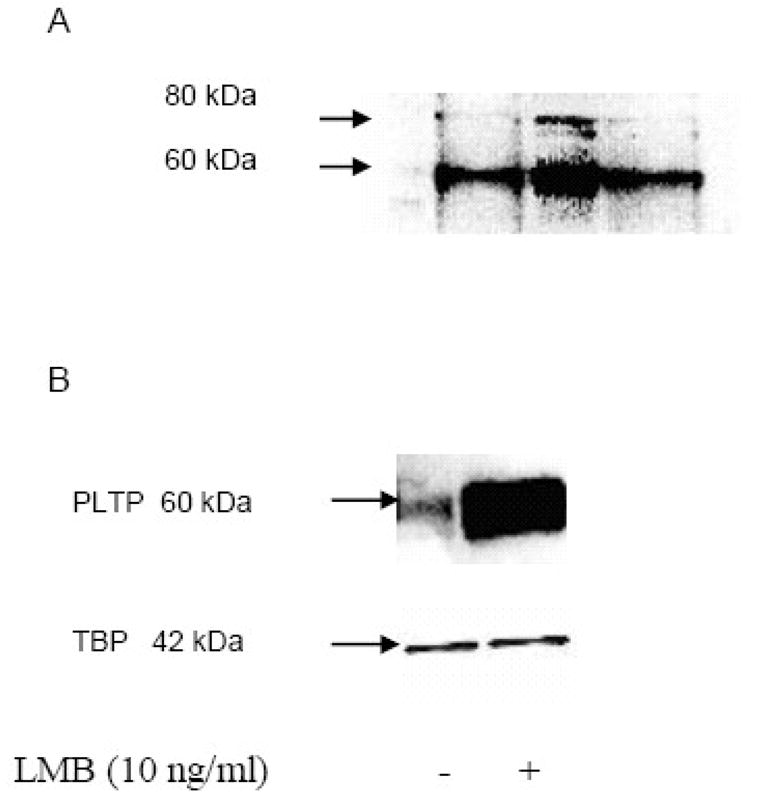
PLTP is present in nuclear protein extracts of SK-N-SH neuroblastoma and human cortical neurons, HCN2 cells, and its nuclear export is chromosome region maintenance 1 (CRM1) dependent. A) Isolated SK-N-SH nuclear proteins from three different samples were resolved using SDS-PAGE, and visualized using Western blot analysis with monoclonal anti-PLTP antibody. B) PLTP Western blot analysis of nuclear proteins isolated from HCN2 cells incubated without (−) or with (+) 10 ng/ml LMB, a specific inhibitor of nuclear export mediated by CRM1, for 24 hours under serum-free conditions. PLTP was identified using monoclonal anti-PLTP antibody. Loading control was evaluated using anti-TATA binding protein (TBP). Secondary antibody controls were negative (not shown).
To evaluate whether PLTP accumulates in nuclei in presence of the chromosome region maintenance 1 (CRM1) protein, the main nuclear export protein, we incubated neuronal cells with leptomycin B (LMB), a specific inhibitor of CRM1 [22–24]. HCN2 cells were incubated with LMB, and isolated nuclear proteins were evaluated by Western blot analyses (Figure 1b). Similar results were obtained using neuroblastoma (SK-N-SH) cells (not shown). Additionally, we evaluated how glycosylation affects nuclear accumulation of PLTP. Removal of carbohydrate from four N-glycosylation sites of PLTP, using site-directed mutagenesis, increased the amount of PLTP in the nuclei of transfected CHO cells, and its retention in the presence of LMB (Figure 2).
Figure 2.
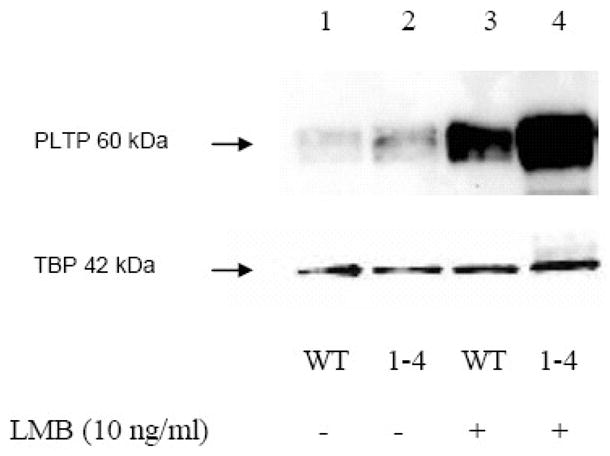
PLTP Western blot analysis of nuclear proteins from CHO cells transfected with the wild-type PLTP (lanes 1 and 3) or PLTP mutant for four glycosylation sites (lanes 2 and 4), incubated without (lanes 1 and 2), or with leptomycin B (LMB; lanes 3 and 4). PLTP was identified using monoclonal anti-PLTP antibody. Loading control was evaluated using anti-TATA binding protein (TBP). Secondary antibody controls were negative (not shown).
We also performed immunocytochemistry on cells incubated with or without LMB, using monoclonal antibody against PLTP. Our analyses confirmed that incubation of SK-N-SH and HCN2 cells with LMB leads to accumulation of PLTP in the nuclei (Figures 3 and 4). To confirm that accumulated protein is indeed PLTP, we incubated mock-transfected and PLTP-EGFP transfected BHK cells with LMB. One hour after the addition of LMB, we observed live cells under the fluorescent microscope to evaluate localization of PLTP-EGFP (Figure 5a). At the end of the experiment, fixed cells transfected with PLTP-EGFP were also observed under confocal microscope to confirm intranuclear localization of PLTP-EGFP (Figure 5b-h). Both fluorescent and confocal microscopy confirmed that PLTP is present in the nuclei of the transfected cells, and that it accumulates in nuclei of cells incubated with LMB.
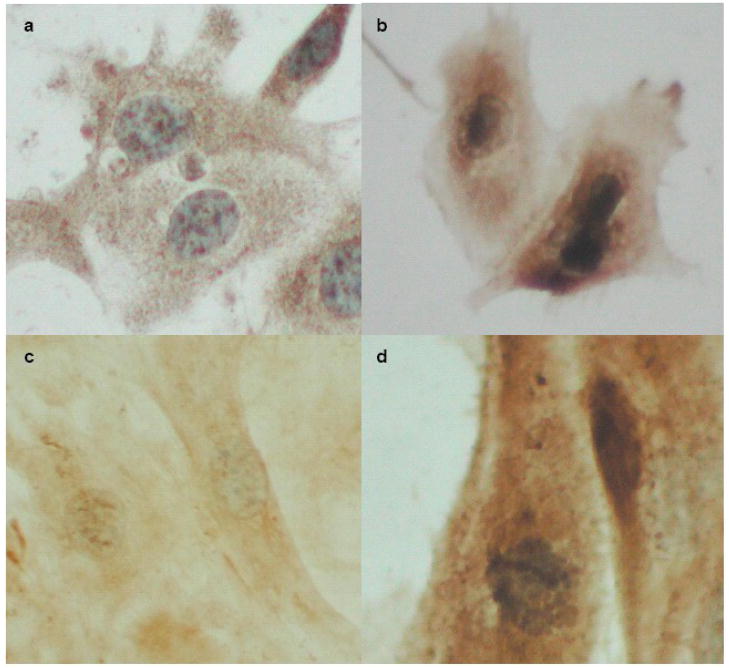
Figure 3.
PLTP immunocytochemistry in neuronal cells. SK-N-SH cells incubated without (a) or with (b) leptomycin B (LMB) for 24 hours were stained with monoclonal antibody against PLTP, and visualized using DAB (brown) and counterstained with hematoxylin (blue). Human cortical neurons (HCN2 cells) incubated without (c) or with 10 ng/ml LMB for 24 hours (d) are shown for comparison. Micrographs were obtained using Olympus microscope and Nikon E990 digital camera.
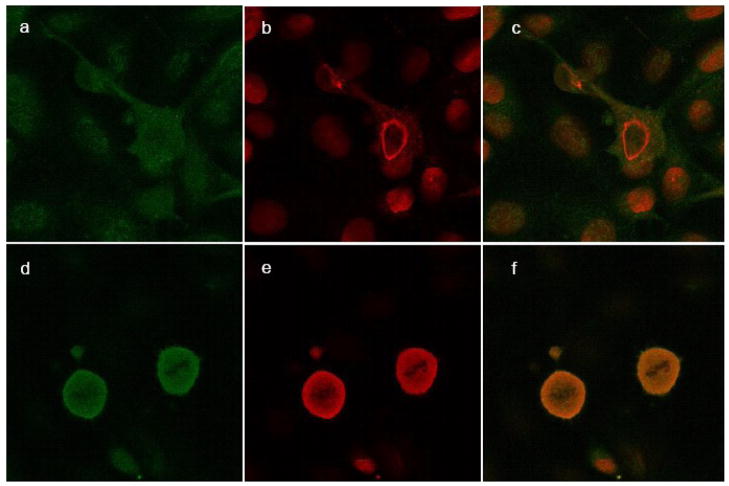
Figure 4.
Confocal microscopy of nuclear PLTP in SK-N-SH cells. SK-N-SH cells incubated without (a–c) and with LMB (d–f) for 24 hours were stained with monoclonal antibody against PLTP (green; a and d) and Lamin A, a nuclear marker (red; b and e); merged files (c and f). Secondary antibody controls were negative (not shown). The slides were observed under confocal Zeiss LSM 510 Meta microscope, using a water-immersion objective.
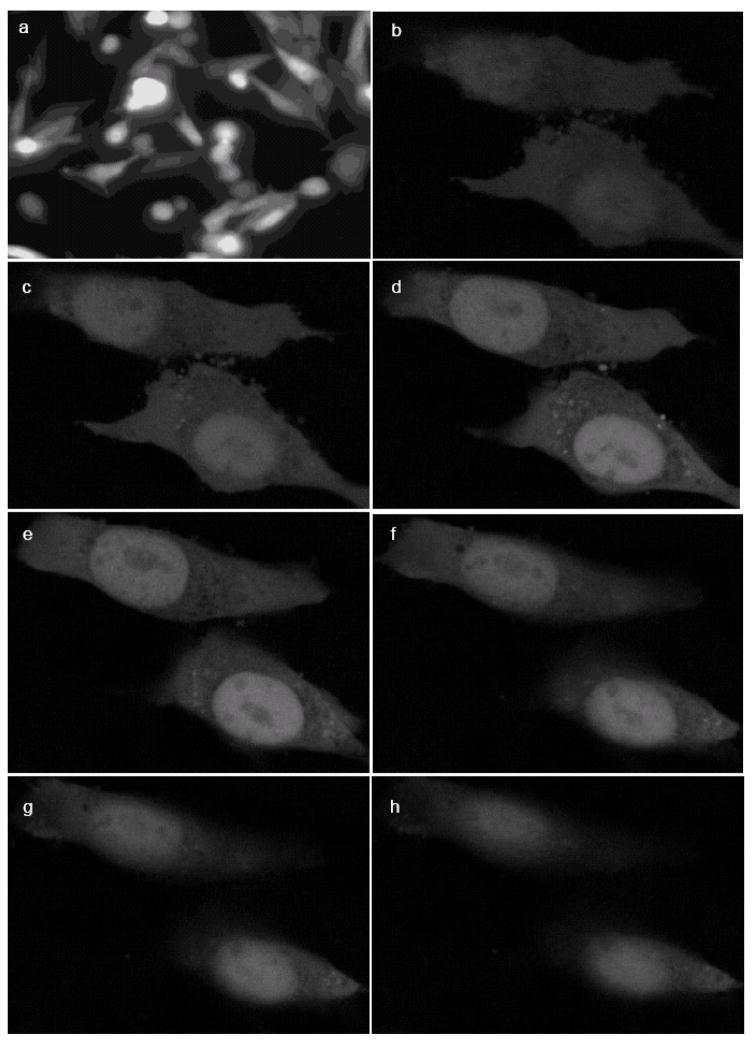
Figure 5.
EGFP-tagged PLTP accumulates in the nuclei of BHK cells incubated with leptomycin B (LMB). Image of live PLTP-EGFP transfected (a) BHK cells incubated with leptomycin B, viewed under inverted fluorescent microscope after one hour of incubation with LMB, and representative serial confocal microscopy images (Z-stack) of formaldehyde-fixed PLTP-EGFP transfected BHK cells following 24h incubation with LMB (b–h).
In order to establish whether secreted PLTP can enter the cell and nucleus, conditioned media from control BHK cells and media of BHK cells transfected with PLTP-EGFP were incubated with the control BHK cells for 24 hours in the presence or absence of LMB, and observed under confocal microscope. Cells incubated with PLTP-EGFP containing conditioned media without LMB showed sparse positive intracellular fluorescence, with approximately 1% of cells reaching fluorescence threshold necessary for observation (not shown). Cells incubated with PLTP-EGFP containing conditioned media with addition of LMB showed much higher percentage of clearly positive cells, with majority of cells containing faint fluorescence, and approximately 20% of cells reaching medium or high intracellular fluorescence levels (Figure 6). Positive cells incubated with LMB showed clear accumulation of fluorescence in the nuclei. Control cells, incubated with conditioned media from non-transfected BHK cells in the presence or absence of LMB were negative (not shown).
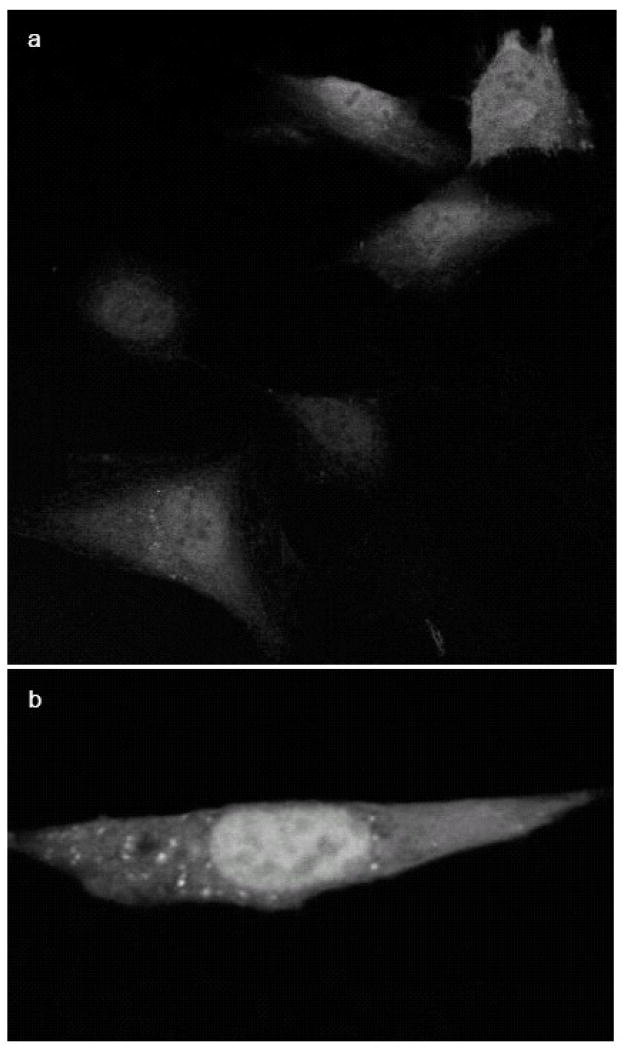
Figure 6.
PLTP-EGFP re-enters the cells and translocates to the nuclei. Non-transfected BHK cells were incubated for 24 hours with concentrated conditioned media from cells expressing PLTP-EGFP without (not shown) and with 10 ng/ml leptomycin B (a and b). Control cells incubated with media from non-transfected BHK cells, without or with LMB, were negative (not shown).
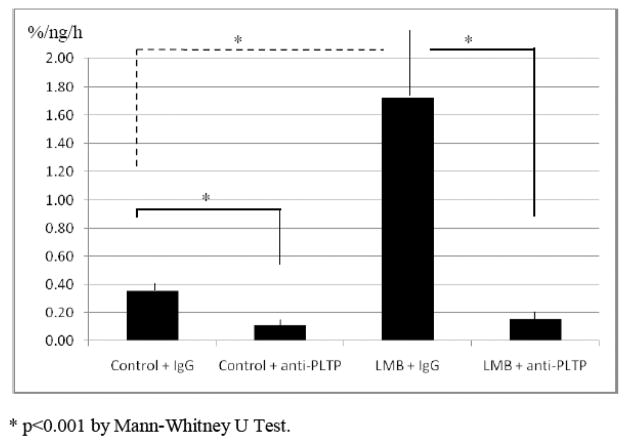
To establish whether nuclear PLTP is active in phospholipid transfer, we performed an in vitro phospholipid transfer assay using nuclear extract from HCN2 cells. Intranuclear PLTP was active in phospholipid transfer and PLTP-mediated phospholipid transfer activity was significantly increased in cells incubated with LMB (control 0.36 ± 0.05 % transfer/ng nuclear protein/h vs. LMB 1.72 ± 0.43 %/ng/h). Incubation with specific chicken polyclonal anti-PLTP antibody significantly reduced measured intranuclear phospholipid transfer activity (control with anti-PLTP antibody 0.11 ± 0.05 %/ng/h, and LMB with anti-PLTP antibody 0.16 ± 0.06 %/ng/h; Figure 7).
Figure 7.
Intranuclear PLTP-mediated phospholipid transfer activity in HCN2 cells. Nuclear extracts of HCN2 cells incubated without (control) or with LMB (10 ng/ml) for 24 hours were analyzed for PLTP-mediated phospholipid transfer activity before and after incubation with chicken polyclonal anti PLTP antibody. Transfer of radioactively-labeled (C14) phospholipid from liposome to HDL particles was measured in vitro. Activity in phospholipid transfer is expressed as percent radioactively labeled phospholipid transferred to the acceptor per nanogram of total nuclear protein per hour (%/ng/h).
We also conducted computer analysis of the PLTP protein sequence for the presence of known nuclear motifs involved in nuclear import and export. The classical nuclear localization signal (NLS) contains basic amino-acids (R = arginine, K = lysine, and H = histidine) in a monopartite sequence [K(K/R)XK(K/R)], which typically requires the presence of lysine residue at the first position, or in a bipartite sequence that consists of two groups of basic amino-acids separated by 10–12 amino-acid linker region [17–20]. Using an online tool for search of the consensus nuclear localization sequences (see Methods), we could not establish that PLTP contains a consensus sequence for nuclear import, either the canonical or the bi-partite sequence. This finding is not unusual, as nuclear import sequences are more heterogeneous, although most of them require presence of a cluster of basic amino-acids. Two regions fulfilling this later requirement were identified in PLTP, one in the carboxyl-terminal and one in the amino-terminal barrel of PLTP (Table 1). In addition, a newly identified NLS motif (S/T-P-S/T), which responds to extracellular stimuli and requires phosphorylation for transfer into the nucleus [25], is present at the C-terminal of PLTP (Table 1).
Table 1.
Putative nuclear localization (NLS) and export signal (NES) motif candidates in PLTP molecule (numbering is based on PLTP protein sequence containing signal peptide).
| PLTP sequence | Putative NLS motif/region |
|---|---|
| a) 100–104 | RFRRQ |
| b) 372–400 | |
| RLSAKMALRGKALRTQLDLRRFRIYSNH | |
| c) 487–489 | TPS |
| PLTP sequence | Putative NES motif |
|
| |
| 1) 3–12 | LFGALFLALL |
| 2) 42–51 | LEGCLETITI |
| 3) 68–75 | VKVTELQL |
| 4) 88–99 | LMLQITNASLGL |
| 5) 123–131 | VSIRTGLEL |
| 6) 164–180 | VYDFLSTFITSGMRFLL |
| 7) 288–302 | LLLVGDKVPHDLDML |
| 8) 311–327 | IVLLSPAVIDSPLKLEL |
| 9) 344–355 | ISVTASVTIALV |
| 10) 401–419 | LESLALIPLQAPLKTMLQI |
Furthermore, we identified several potential consensus nuclear export sequence (NES) motifs (consensus sequence: ΦX(2–3) ΦX(2–3) ΦXΦ where Φ stands for either leucine, L, isoleucine, I, valine, V, phenylalanine, F, or methionine, M; while X stands for any aminoacid) [21]. Putative consensus NES sequences are distributed in both the amino- and carboxyl-terminal portion of the PLTP molecule (Table 1). Further evaluation by the online tool, NetNES 1.1, suggested that each of the identified putative NES motifs fulfills the basic requirements for nuclear export. Functionality of these motifs remains to be established.
Discussion
In the current study, we present evidence for PLTP localization in the nucleus of neuronal cells, as well as in mammalian ovary and kidney cells transfected with human PLTP. These findings indicate that PLTP is present in the nuclei of different cell types, and its export from the nucleus is CRM1-dependent. We also provide evidence for reentry of extracellular PLTP into the cell and translocation into the nucleus. Furthermore, the assay of PLTP phospholipid transfer activity before and after incubation with anti-PLTP antibody confirmed that intranuclear PLTP is active in phospholipid transfer.
The current paradigm for protein localization implies that proteins destined for secretion are targeted to the endoplasmic reticulum (ER) by a signal peptide sequence at the beginning of mRNA transcription, then transferred to Golgi and secreted into extracellular space. It is, therefore, unclear by which mechanism PLTP enters the nucleus. One possibility is that the signal peptide coded by Exon 2 in PLTP may be omitted through alternative splicing, or masked by some other mechanism. An alternative explanation would be that secreted PLTP re-enters the cells, escapes the endocytic pathway and enters the nucleus. This type of nuclear translocation is rare, but has been reported. For example, apoE, a molecular partner of PLTP, has been shown to translocate into the nucleus following entry into cells through an apoE receptor-dependent mechanism [26]. Furthermore, apolipoprotein D has been shown to translocate into the nucleus [27]. One potential explanation may be in the fact that PLTP, apoE and apoD are bound to lipoprotein particles, which could be taken up by the cells as holoparticles, allowing for intracellular and intranuclear localization and function. Indeed, a recently published study has shown that only apoE associated with lipids is capable of nuclear translocation [26]. These findings raise an interesting possibility that not only cytoplasmic, but also extracellular lipids can enter the nucleus, providing an important signaling mechanism through which astrocytes, for example, may be affecting neuronal nuclear-based functions associated with lipids. Our study provides evidence that at least a portion of secreted PLTP can re-enter the cell, and that PLTP can translocate to the nucleus upon re-entry. Interestingly, cells incubated with secreted PLTP-EGFP show variable intracellular fluorescence, suggesting the possibility that PLTP reuptake is regulated by some cellular parameter, such as the cell cycle stage.
It is also possible that the current cellular secretory pathway paradigm is not correct or that some sequence motif in a target protein may allow for exit from the secretory pathway and consequently permit intracellular or intranuclear localization. For example, numerous recent studies suggest involvement of the endoplasmic reticulum quality control system, ERAD, as a putative mechanism for transfer of proteins from the ER to cytoplasm and even nucleus [28–33]. ERAD is typically involved in transfer of misfolded proteins from the ER for degradation by the ubiquitin system via an ER translocon, sec61. Recently published studies suggest that this system is also involved in transfer of secreted proteins to cytoplasmic and nuclear compartments [28–33]. An interesting review by Arnoys and Wang lists nearly eighty well-documented secreted proteins that are localized in the nucleus [34]. Data from our experiments suggest that PLTP re-entry into the cell only partially explains PLTP presence in the nucleus, while additional nuclear PLTP likely comes from the intracellular pool that did not get secreted. Therefore, it would be important to further explore putative mechanisms involved in the intracellular and intranuclear localization of PLTP.
Although it is currently not known what functional roles PLTP may be playing in the nucleus, there are several likely possibilities. Typically, shuttling of proteins between cytoplasm and nucleus is important for regulation of cellular functions, such as cell proliferation and differentiation, and for maintenance of the cellular structural stability and function. Alterations in nucleocytoplasmic shuttling for numerous transcription factors have been associated with pathophysiological processes involved in development of AD, Parkinson’s disease, amyotrophic lateral sclerosis and Huntington’s disease, among others (reviewed in [35]). Gene expression can be regulated by factors that modify chromatin and by signaling molecules, as well as by factors regulating lipid content of the nucleus [35–37].
We have shown that intranuclear PLTP is active in lipid transport, suggesting that PLTP may be involved in regulation, transport and distribution of phospholipids and cholesterol in the nucleus. Numerous studies have shown that lipids are an integral part of the nucleus [38–43]. Lipids are present not only in the nuclear envelope, but also in the nuclear matrix and chromatin [37, 44–45]. Additionally, elegant experiments by Albi and colleagues have shown that at least some of the phospholipids present in chromatin, which represent approximately 10% of the nuclear phospholipids, are transported into the nucleus from the cytoplasm [46], while others may be synthesized in situ [37]. Although endonuclear lipids were initially considered relevant only for structural stability of the nucleus, an increasing body of evidence suggests their functional contribution to other nuclear processes. For example, endonuclear lipids have been associated with endonuclear lipid second messenger signaling, thus affecting processes involved in cell proliferation, differentiation and apoptosis [36, 37]. Furthermore, nuclear composition of lipids is cell type and cell cycle phase specific [37]. Despite the overwhelming evidence of lipid transfer of cholesterol and phospholipids to and from the nucleus, the processes and components of this lipid transfer are currently unknown [37]. Given the PLTP’s wide affinity for free cholesterol and different classes of phospholipids, it is reasonable to speculate that PLTP may be involved in this process.
PLTP is also necessary for transfer of α-tocopherol to cells and tissues [11]. Guarnieri and colleagues have shown that α-tocopherol is present in the nuclei, and suggested that α-tocopherol may be involved in the regulation of RNA synthesis [47]. Further studies confirmed the importance of α-tocopherol in preservation of RNA structure, and reduction in nuclear α-tocopherol content had similar effects on nuclear components as aging [48]. Additionally, intranuclear antioxidative potential may be relevant not only in aging, but also in pathophysiology of various diseases, such as Alzheimer’s disease or cancer. Since PLTP is critical for transfer of α-tocopherol to the cells, it is possible that it may be also involved in its transfer to the nucleus.
A recent study implicated PLTP as the main component in one of the nine pathways that regulate survival in breast cancer [49], and PLTP has been differentially expressed in numerous tumors, including different types of leukemia ([50–52] and data obtained through data mining), but the mechanisms through which PLTP may be affecting these processes are currently unknown. It is possible that intracellular, and in particular intranuclear PLTP localization and functions are relevant for these processes, and unraveling the mechanisms controlling PLTP intracellular compartmentalization would be important. Our study and its potential implications may be relevant for our understanding of the roles of PLTP in numerous human diseases, including Alzheimer’s disease, cardiovascular disease, diabetes and cancer.
Acknowledgments
We are grateful to Dr. Vesa Olkkonen from the National Public Health Institute, Helsinki, Finland, for generous gift of control and PLTP-EGFP vectors. We are thankful to Dr. Kevin O’Brien for use of digital imaging system; Drs. Charles Murry and Hans Reinecke for use of Zeiss fluorescent microscope; Dr Ronald Seifert from the University of Washington South Lake Union confocal microscopy facility for his help and training; and to Hal Kennedy for his help with manuscript preparation.
Funding
This study was supported by the NIH NHLBI grant HL030086.
Footnotes
Conflict of Interest
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Day JR, Albers JJ, Lofton-Day CE, Gilbert TL, Ching AF, Grant FJ, O’Hara PJ, Marcovina SM, Adolphson JL. Complete cDNA encoding human phospholipid transfer protein from human endothelial cells. J Biol Chem. 1994;269:9388–9391. [PubMed] [Google Scholar]
- 2.Cheung MC, Albers JJ. Active plasma phospholipid transfer protein is associated with apoA-I but not apoE-containing lipoproteins. J Lipid Res. 2006;47:1315–1321. doi: 10.1194/jlr.M600042-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Vuletic S, Peskind ER, Marcovina SM, Quinn JF, Cheung MC, Kennedy H, Kaye JA, Jin LW, Albers JJ. Reduced CSF PLTP activity in Alzheimer’s disease and other neurologic diseases; PLTP induces ApoE secretion in primary human astrocytes in vitro. J Neurosci Res. 2005;80:406–13. doi: 10.1002/jnr.20458. [DOI] [PubMed] [Google Scholar]
- 4.Albers JJ, Cheung MC. Emerging roles for phospholipid transfer protein in lipid and lipoprotein metabolism. Curr Opin Lipidol. 2004;15:255–260. doi: 10.1097/00041433-200406000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Albers JJ, Wolfbauer G, Cheung MC, Day JR, Ching AF, Lok S, Tu AY. Functional expression of human and mouse plasma phospholipid transfer protein: effect of recombinant and plasma PLTP on HDL subspecies. Biochim Biophys Acta. 1995;1258:27–34. doi: 10.1016/0005-2760(95)00091-p. [DOI] [PubMed] [Google Scholar]
- 6.Nishida HI, Nishida T. Phospholipid transfer protein mediates transfer of not only phosphatidylcholine but also cholesterol from phosphatidylcholine-cholesterol vesicles to high density lipoproteins. J Biol Chem. 1997;272:6959–6964. doi: 10.1074/jbc.272.11.6959. [DOI] [PubMed] [Google Scholar]
- 7.Rao R, Albers JJ, Wolfbauer G, Pownall HJ. Molecular and macromolecular specificity of human plasma phospholipid transfer protein. Biochemistry. 1997;36:3645–3653. doi: 10.1021/bi962776b. [DOI] [PubMed] [Google Scholar]
- 8.Kostner GM, Oettl M, Jauhiainen M, Ehnholm C, Esterbauer H, Dieplinger H. Human plasma phospholipid transfer protein accelerates exchange/transfer of alpha-tocopherol between lipoproteins and cells. Biochem J. 1995;305:659–67. doi: 10.1042/bj3050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagrost L, Desrumaux C, Masson D, Deckert V, Gambert P. Structure and function of the plasma phospholipid transfer protein. Curr Opin Lipidol. 1998;9:203–209. doi: 10.1097/00041433-199806000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Qin S, Kawano K, Bruce C, Lin M, Bisgaier C, Tall AR, Jiang XC. Phospholipid transfer protein gene knockout mice have low high density lipoprotein levels, due to hypercatabolism, and accumulate apoA-IV-rich lamellar lipoproteins. J Lipid Res. 2000;41:269–276. [PubMed] [Google Scholar]
- 11.Desrumaux C, Risold PY, Schroeder H, Deckert V, Masson D, Athias A, Laplanche H, Guern NL, Blache D, Jiang XC, Tall AR, Desor D, Lagrost L. Phospholipid transfer protein (PLTP) deficiency reduces brain vitamin E content and increases anxiety in mice. FASEB J. 2005;19:296–297. doi: 10.1096/fj.04-2400fje. [DOI] [PubMed] [Google Scholar]
- 12.Vuletic S, Jin LW, Marcovina SM, Peskind ER, Moller T, Albers JJ. Widespread distribution of PLTP in human CNS: evidence for PLTP synthesis by glia and neurons, and increased levels in Alzheimer’s disease. J Lipid Res. 2003;44:1113–23. doi: 10.1194/jlr.M300046-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Arai K, Ikegaya Y, Nakatani Y, Kudo I, Nishiyama N, Matsuki N. Phospholipase A2 mediates ischemic injury in the hippocampus: a regional difference of neuronal vulnerability. Eur J Neurosci. 2001;13:2319–2323. doi: 10.1046/j.0953-816x.2001.01623.x. [DOI] [PubMed] [Google Scholar]
- 14.Siggins S, Ehnholm C, Jauhiainen M, Olkkonen VM. Plasma phospholipid transfer protein fused with green fluorescent protein is secreted by HepG2 cells and displays phosphatidylcholine transfer activity. Biochem Cell Biol. 2006;84:117–125. doi: 10.1139/o05-168. [DOI] [PubMed] [Google Scholar]
- 15.Cheung MC, Wolfbauer G, Albers JJ. Plasma phospholipid mass transfer rate: relationship to plasma phospholipid and cholesteryl ester transfer activities and lipid parameters. Biochim Biophys Acta. 1996;1303:103–10. doi: 10.1016/0005-2760(96)00082-3. [DOI] [PubMed] [Google Scholar]
- 16.Cheung MC, Wolfbauer G, Deguchi H, Fernandez JA, Griffin JH, Albers JJ. Human plasma transfer protein specific activity is correlated with HDL size; implications for lipoprotein physiology. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbalip.2008.12.010. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 18.Damelin M, Silver PA, Corbett AH. Nuclear protein transport. Methods Enzymol. 2002;351:587–607. doi: 10.1016/s0076-6879(02)51870-x. [DOI] [PubMed] [Google Scholar]
- 19.Xiao Z, Latek R, Lodish HF. An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity. Oncogene. 2003;22:1057–1069. doi: 10.1038/sj.onc.1206212. [DOI] [PubMed] [Google Scholar]
- 20.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition. function and interaction with importin. J Biol Chem. 2005;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 23.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 24.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 25.Chuderland D, Konson A, Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol Cell PMID. 2008;18760948 doi: 10.1016/j.molcel.2008.08.007. In Press. [DOI] [PubMed] [Google Scholar]
- 26.Kim WS, Elliot DA, Kockx M, Kritharides L, Rye KA, Jans DA, Garner B. Analysis of apolipoprotein E nuclear localization using green fluorescent protein and biotinylation approaches. Biochem J. 2008;409:701–709. doi: 10.1042/BJ20071261. [DOI] [PubMed] [Google Scholar]
- 27.Do Carmo S, Levros L-C, Rassart E. Modulation of apolipoprotein D expression and translocation under specific stress conditions. Biochim Biophys Acta – Mol Cell Res. 2007;1773:954–969. doi: 10.1016/j.bbamcr.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- 29.Brandizzi F, Hanton S, DaSilva LL, Boevink P, Evans D, Oparka K, Denecke J, Hawes C. ER quality control can lead to retrograte transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 2003;34:269–281. doi: 10.1046/j.1365-313x.2003.01728.x. [DOI] [PubMed] [Google Scholar]
- 30.Carpenter G. Nuclear localization and possible functions of receptor tyrosine kinases. Curr Opin Cell Biol. 2003;15:143–148. doi: 10.1016/s0955-0674(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 31.Johnson HM, Subramaniam PS, Olsnes S, Jans DA. Trafficking and signaling pathways of nuclear localizing protein ligands and their receptors. BioEssays. 2004;26:993–1004. doi: 10.1002/bies.20086. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q, Lianyun L, Yihong Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Cell Biol. 2006;174:963–971. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao HJ, Carpenter G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol Biol Cell. 2007;18:1064–1072. doi: 10.1091/mbc.E06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnoys EJ, Wang JL. Dual localization: proteins in extracellular and intracellular compartments. Acta Histochem. 2007;109:89–110. doi: 10.1016/j.acthis.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Chu CT, Plowey ED, Want Y, Patel V, Jordan-Sciutto KL. Location, location, location: altered transcription factor trafficking in neurodegeneration. J Neuropathol Exp Neurol. 2007;66:873–883. doi: 10.1097/nen.0b013e318156a3d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albi E, Viola Magni MP. The role of intranuclear lipids. Biol Cell. 2004;96:657–667. doi: 10.1016/j.biolcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Hunt AN. Dynamic lipidomics of the nucleus. J Cell Biochem. 2006;97:244–251. doi: 10.1002/jcb.20691. [DOI] [PubMed] [Google Scholar]
- 38.La Cour LF, Chayen J, Gahan PB. Evidence for lipid material in chromosomes. Exp Cell Res. 1958;14:469–485. doi: 10.1016/0014-4827(58)90155-1. [DOI] [PubMed] [Google Scholar]
- 39.Gahan PB. Histochemical evidence for the presence of lipids on the chormosomes of animal cells. Exp Cell Res. 1965;39:136–144. doi: 10.1016/0014-4827(65)90016-9. [DOI] [PubMed] [Google Scholar]
- 40.Spangler M, Coetzee ML, Katlyl SL, Morris HP, Ove P. Some biochemical characteristics of rat liver and Morris hepatoma nuclei and nuclear membranes. Cancer Res. 1975;35:3145. [PubMed] [Google Scholar]
- 41.Upreti GC, De Autmno RJ, Wood R. Membrane lipids of hepatic tissue II. Phospholipids from subcellular fractions of liver and hepatoma 7288CTC. J Natl Cancer Inst. 1983;70:567–573. [PubMed] [Google Scholar]
- 42.Song M, Rebel G. Rat liver nuclear lipids. Composition and biosynthesis. Basic Appl Histochem. 1987;31:377–387. [PubMed] [Google Scholar]
- 43.Albi E, Viola Magni MP. The presence and the role of chromatin cholesterol in rat liver regeneration. J Hepatol. 2002;36:395–400. doi: 10.1016/s0168-8278(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 44.Rose HG, Frenster JH. Composition and metabolism of lipids within repressed and active chromatin in interphase lymphocytes. Biochim Biophys Acta. 1965;106:577–591. doi: 10.1016/0005-2760(65)90073-1. [DOI] [PubMed] [Google Scholar]
- 45.Cocco L, Maraldi NM, Manzoli FA, Gilmour RS, Lang A. Phospholipid interactions in rat liver nuclear matrix. Biochem Biophys Res Commun. 1980;96:890–898. doi: 10.1016/0006-291x(80)91439-4. [DOI] [PubMed] [Google Scholar]
- 46.Albi E, Mersel M, LeRay C, Tomassoni ML, Viola Magni MP. Rat liver chromatin phospholipids. Lipids. 1994;29:715–719. doi: 10.1007/BF02538916. [DOI] [PubMed] [Google Scholar]
- 47.Guarnieri C, Flamigni F, Caldarera CR. Subcellular localization of alpha-tocopherol and its effect on RNA synthesis in perfused rabbit heart. Ital J Biochem. 1980;29:176–184. [PubMed] [Google Scholar]
- 48.Malatesta M, Bertoni-Freddari C, Fattoretti P, Caporaloni C, Fakan S, Gazzanelli G. Altered RNA structural constituents in aging and vitamin E deficiency. Mech Ageing Dev. 2003;124:175–181. doi: 10.1016/s0047-6374(02)00117-3. [DOI] [PubMed] [Google Scholar]
- 49.Ferkingstad E, Frigessi A, Lyng H. Indirect genomic effects on survival from gene expression data. Genome Biol. 2008;9:R58–R72. doi: 10.1186/gb-2008-9-3-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skamrov AV, Nechaenko MA, Goryunova LE, Feoktistova ES, Khaspekov GL, Kovalevsky DA, Vinnitsky LI, Sheremeteva GF, Beabealashwilli RS. Gene expression analysis to identify mRNA markers of cardiac myxoma. J Mol Cell Cardiol. 2004;37:717–733. doi: 10.1016/j.yjmcc.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 51.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimndi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui CH, Evans WE, Naeve C, Wong L, Downing J. Classification, subtype discovery, and prediction of outcome in pediatric lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 52.Shi X, Cao S, Kitsuhashi M, Xiang Z, Ma X. Genome-wide analysis of molecular changes in IL-12-induced control of mammary carcinoma via IFN-γ-independent mechanisms. J Immunol. 2004;172:4111–4122. doi: 10.4049/jimmunol.172.7.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]