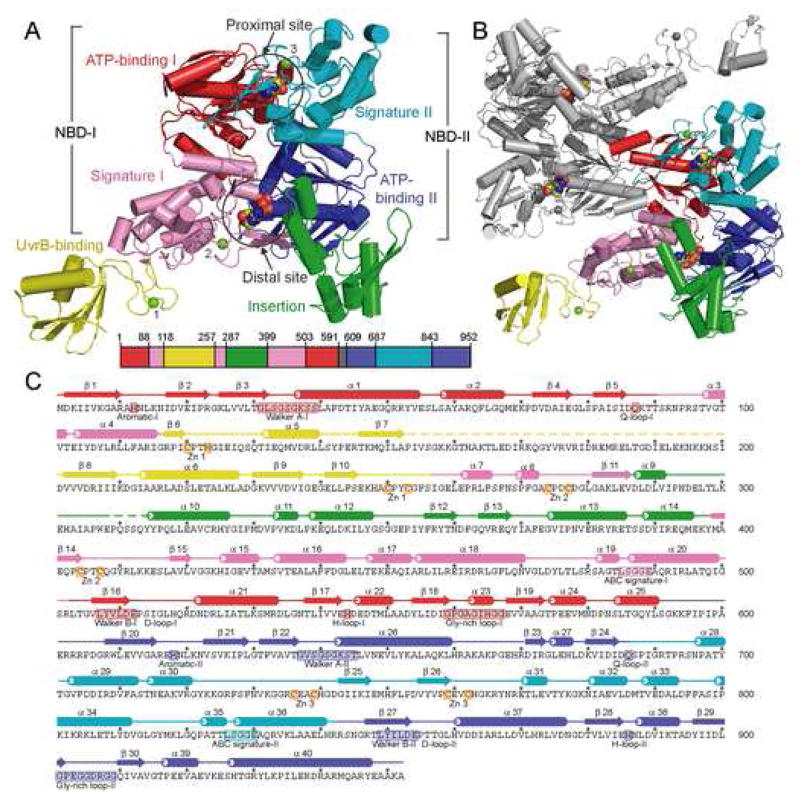
Figure 1.
Structure of Bacillus stearothermophilus UvrA. (A) Overall structure of the UvrA monomer, with a-helices depicted as cylinders and β-strands as arrows. The protein is colored by domains, ATP-binding I (1–87,503–590), red; signature I (88–117, 257–286, 399–502), pink; ATP-binding II (609–686, 843–952), blue; signature II (687–842), cyan; UvrB-binding (118–256), yellow; insertion (287–398), green; and linker (591–608), gray; with the Zn atoms, numbered by module, in light green. The bound ADP molecules are shown as space-filling models. The location of each domain, colored as above, is projected onto the primary sequence of BstUvrA, shown as a bar. (B) Overall structure of the UvrA dimer as observed in the crystal asymmetric unit. One protomer is colored as in (A), and the second protomer in gray. (C) Secondary structure assignment of BstUvrA, colored by domains as above. Disordered regions are depicted as dashed lines. Locations of the conserved ABC ATPase motifs, glycine-rich loops, and Zn-coordinating residues are depicted on the amino acid sequence.