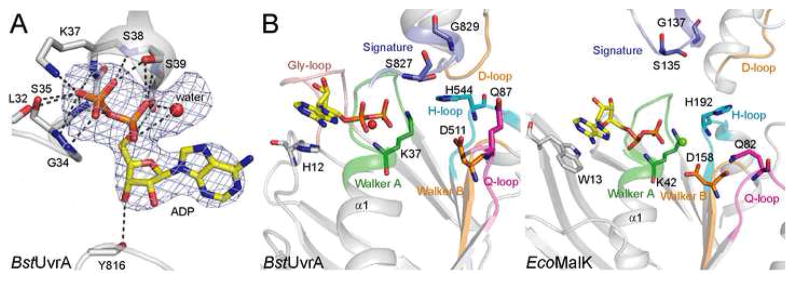
Figure 3.
Nucleotide binding by BstUvrA. (A) Binding of ADP by the Walker A motif. ADP and the interacting protein residues are shown as sticks, with the unknown solvent component, modeled as a water molecule, depicted as a red sphere. Hydrogen bonds are drawn as dashed lines. The difference electron density calculated with the nucleotide and water molecule omitted from the model is shown at 3σ. (B) Comparison of the nucleotide-binding sites from UvrA (left) and MalK (right, PDB code 2AWO). The ATPase motifs are colored as in Figure 2. Important conserved residues are shown as sticks. The conserved glycine-rich loop found in UvrA is peach. All four nucleotide-binding sites in the UvrA dimer are structurally very similar, thus only the proximal site of protomer A is shown.