Abstract
We conducted a genome-wide association study to identify novel genes influencing diastolic blood pressure (BP) response to hydrochlorothiazide, a commonly prescribed thiazide diuretic preferred for the treatment of high BP. Affymetrix GeneChip Human Mapping 100K Arrays were used to measure single nucleotide polymorphisms across the 22 autosomes in 194 non-Hispanic black subjects and 195 non-Hispanic white subjects with essential hypertension selected from opposite tertiles of the race- and sex-specific distributions of age-adjusted diastolic BP response to hydrochlorothiazide (25 mg daily, PO, for 4 weeks). The black sample consisted of 97 “good” responders (diastolic BP response [mean±SD]=-18.3±4.2 mm Hg; age=47.1±6.1 years; 51.5% women) and 97 “poor” responders (diastolic BP response=-0.18±4.3; age=47.4±6.5 years; 51.5% women). Haplotype trend regression identified a region of chromosome 12q15 in which haplotypes constructed from 3 successive single nucleotide polymorphisms (rs317689, rs315135, and rs7297610) in proximity to lysozyme (LYZ), YEATS domain containing 4 (YEATS4), and fibroblast growth receptor substrate 2 (FRS2) were significantly associated with diastolic BP response (nominal P=2.39×10-7; Bonferroni corrected P=0.024; simulated experiment-wise P=0.040). Genotyping of 35 additional single nucleotide polymorphisms selected to “tag” linkage disequilibrium blocks in these genes provided corroboration that variation in LYZ and YEATS4 was associated with diastolic BP response in a statistically independent data set of 291 black subjects and in the sample of 294 white subjects. These results support the use of genome-wide association analyses to identify novel genes influencing antihypertensive drug responses.
Keywords: hypertension, pharmacogenetics, diuretic, blood pressure, genome
Despite the availability of multiple antihypertensive drugs, acting on a variety of blood pressure (BP)-regulating systems, <40% of treated patients achieve BP control.1 For agents from each class of antihypertensive drugs, BP responses vary among patients, with SDs of responses as large as mean responses, and ranges of responses several times greater than mean responses.2 This variation appears to be the result of pharmacodynamic differences in drug action rather than pharmacokinetic differences in drug levels and likely reflects the heterogeneity of pathophysiologic mechanisms contributing to hypertension.3 Knowledge of genetic variants that influence BP responses has the potential to improve on the customary trial-and-error approach to antihypertensive therapy.4
Thiazide diuretics are recommended as initial and preferred therapy in most patients,5 but when used as monotherapy, BP is controlled in only ≈50% of patients.6 Established predictors of greater BP responses to diuretics include black race, older age, and lower activity of the renin-angiotensin-aldosterone system.7 Measurements of polymorphisms in candidate genes encoding components of the renin-angiotensin-aldosterone system or the renal sodium transport systems targeted by diuretics have been reported to improve the ability to predict antihypertensive responses.8,9 However, after all of the identified predictors were considered in statistical models, the majority of interindividual variation in BP responses remained unexplained.8 To advance beyond the limitations of assessing only polymorphisms in known or suspected candidate genes, we have undertaken a large-scale genomic association study to identify novel genes influencing antihypertensive drug responses.
Methods
Phenotypic data and biological samples were collected as part of the Genetic Epidemiology of Responses to Antihypertensives Study conducted between 1997 and 2002.10 The initial objective was to determine whether polymorphisms in candidate genes encoding components of the renin-angiotensin-aldosterone system were predictive of the BP response to the thiazide diuretic, hydrochlorothiazide, in hypertensive non-Hispanic black subjects from Atlanta (n=300) and non-Hispanic white subjects from Rochester (n=300). A standardized study protocol was conducted in the Centers for Transitional Science Activities of Emory University and the Mayo Clinic, in which volunteers 30.0 to 59.9 years of age were instructed to adhere to a standard dietary sodium intake (1 mmol/kg of body weight) and to discontinue previous antihypertensive medications for ≥4 weeks. Once stable elevation of the BP was achieved, a standard dose of hydrochlorothiazide (25 mg/d) was administered orally for 4 weeks. At the end of the drug-free and drug-treatment periods, 3 readings of BP were made by a trained assistant using a random 0 sphygmomanometer after the participant had been seated quietly for ≥5 minutes. The difference between averages of the second and third diastolic BP readings taken before and after hydrochlorothiazide was calculated as the primary measure of drug response.10
To maximize statistical power, a case-control study was designed to contrast good responders (case subjects) with poor responders (control subjects) sampled from opposite tertiles of the distribution of diastolic BP response. Diastolic rather than systolic BP was used to categorize response, because it was the primary measure used to qualify subjects and measure responses to drug therapy in the parent study.10 Before selecting the contrasting groups, the race- and gender-specific distributions of diastolic BP response were adjusted to remove variation attributable to differences in age and pretreatment level of BP, which were found previously to be predictors of BP response to hydrochlorothiazide.10 Within each race and gender stratum, the 50 “best” and 50 “poorest” responders were selected from opposite tertiles of the respective distributions of adjusted diastolic BP response. Separate analyses were planned for each racial group because of anticipated differences in allele frequencies and previous indications of racial differences in predictors of antihypertensive drug responses.10,11
Genotyping
Genomic DNA samples were available from 194 of the black subjects and 195 of the white subjects selected as described above and were genotyped between February 2004 and December 2005 using the GeneChip Human Mapping 100K Array Set, following laboratory protocols recommended by the manufacturer (Affymetrix). Genotyping calls were made by the Dynamic Modeling algorithm12 using a confidence score threshold of 0.25 to filter out potentially erroneous calls as “no-calls.” Analysis of 20 blind duplicate samples from 10 non-Hispanic black subjects and 10 non-Hispanic white subjects indicated a between-assay agreement in genotype calls of 99.7% when averaged over all of the single nucleotide polymorphisms (SNPs).
In a region of chromosome 12q15 identified in non-Hispanic black subjects (see Results section), 35 SNPs were selected for follow-up genotyping within the genes encoding lysozyme (LYZ), YEATS domain containing 4 (YEATS4), and fibroblast growth receptor substrate 2 (FRS2). The SNP selection was designed to provide a high likelihood of capturing a large percentage of common DNA sequence variation across each gene in both racial groups by virtue of tagging blocks of linkage disequilibrium defined by pairwise SNP r2 values at ≥0.8 within the blocks.13 The SNP selection process was completed for each race group separately, and the union of the 2 sets of race-specific tag SNPs was genotyped in both races. For each selected SNP, the relative frequency of the rarer allele was reported to be >0.05 in the HapMap database for Caucasians or the Perlegen database for African Americans.14,15 The tag SNPs were genotyped using the MassARRAY System (Sequenom), which uses PCR amplification followed by an allele-specific ligation reaction, matrix-assisted laser desorption ionization-time-of-flight allele detection, and genotype calling with proprietary software. The sequences of all of the primers and probes are available from the authors on request.
Statistical Analyses
Of the 116 201 SNPs on the Affymetrix GeneChip Human Mapping 100K array set, the number of SNPs suitable for statistical analysis was reduced to 102 334 in black subjects (88.0% of total) and to 95 221 in white subjects (81.9% of total) by the exclusion of SNPs for the following reasons (in order): on the X chromosome (n=2363; 2.0%), monomorphic (in black subjects, n=884, 0.8%; in white subjects, n=7577, 6.5%), minor allele frequency <2% (in black subjects, n=6235, 5.4%; in white subjects, n=7707, 6.6%), relative frequency of no call >20% across samples (in black subjects, n=1273, 1.1%; in white subjects, n=1151, 1.0%), and deviation from Hardy-Weinberg equilibrium at P<0.001 (in black subjects, n=3102, 2.7%; in white subjects, n=2182, 1.9%). Before the pharmacogenomic analyses, we evaluated the black and white samples for evidence of population substructure that might lead to spurious associations.16 Applying the principal component approach implemented in the EIGENSTRAT program to a randomly selected set of 10 000 SNPs from across the genome, we identified no statistically significant evidence of population stratification within either racial group (analyses not shown).
The primary analytic approach to identify pharmacogenomic loci influencing BP response to hydrochlorothiazide was based on analysis of multi-SNP haplotypes rather than single SNP associations. Haplotype trend regression analyses were conducted using HelixTree Genetics Analysis Software version 4.3.0 (Golden Helix), which fits a logistic regression model for categorical outcomes (eg, good versus poor BP response) or a linear regression model for quantitative or ordinal outcomes, and uses a likelihood ratio test for statistical significance.17 The ambiguity of haplotype phase is incorporated into the analysis through use of the posterior probability that a subject has a particular haplotype, as estimated with the expectation-maximization algorithm described by Excoffier and Slatkin.18 For the primary genome-wide analyses, a sliding window of 3 SNPs was used to construct haplotypes and to test for genetic associations with the diastolic BP response category. For the follow-up analyses, comparable analyses of the 3-SNP haplotype sliding windows were conducted using the tag SNPs measured in each positional candidate gene (see Results section), as well as “genecentric” haplotype analyses that used all of the tag SNPs measured in each gene.
To estimate the experiment-wise error for the 3-SNP haplotype associations tested in the primary genome-wide scans, permutation testing was performed by permuting the dependent variable the designated number of times and rerunning the genome-wide haplotype trend regression analysis for each permutation. To compare haplotype frequencies between good and poor responders, we used the score test implemented in the HaploScore program, which addresses limitations resulting from the ambiguity of haplotypes because of the unknown linkage phase of SNPs measured along a chromosome.19
To assess additional evidence to corroborate statistically significant results from the primary haplotype trend regression analysis, we first identified a combination of the tag SNPs, selected as described above, and used it to derive an optimal prediction of diastolic BP response category in the “training sample” data set consisting of categorical diastolic BP response in the 194 black good and poor responders. In lieu of another, separate (ie, replicate) sample from the same population, this prediction model was applied to a statistically independent quantitative measure of residual variation in diastolic BP response in the parent sample of all 291 of the non-Hispanic black subjects, the independence of which was achieved by subtracting off the mean adjusted diastolic BP response of the subgroup to which each subject belonged (good, “intermediate,” or poor). A 1—degree-of-freedom test of haplotype association with BP response was constructed by using the vector of haplotype effect estimates, , from the training data set to derive a linear combination of the expected haplotype effects in the test sample. Specifically, the T statistic:
| (1) |
was calculated, where and are the haplotype parameter vector estimates in the training and test samples, respectively, and V1 is the parameter estimate covariance matrix for the test sample.19 Under the null hypothesis of no haplotype effects, T has an approximately standard normal distribution, uninfluenced by the use of the training sample. When the 2 parameter vectors are proportional, T takes on the value of the square root of the F statistic for the test sample but is subject to only 1 degree of freedom. Furthermore, only the positive tail of the distribution is relevant, and, therefore, a 1-sided test of statistical significance was applied.
For validation in an independent test data set from the white subjects, race specific gene-centric haplotype analyses were applied to the quantitative measure of diastolic BP response in all 294 of the white subjects using a global F test for significance of the association between haplotype variation and BP response. Because of differences in haplotype structure between the races, this approach was preferred over the 1—degree-of-freedom test described above, which was based on application of an optimal model of haplotype effects estimated in the 194 black good and poor responders.
Results
The 194 black and 195 white subjects were, by design, nearly equally divided between good and poor responders and, within each response group, between men and women (Table 1). Also consistent with the sampling design, the good and poor responders differed in mean diastolic BP response (±SD) after adjustment for age and baseline diastolic BP (in black subjects: -18.3±4.2 versus -0.2±4.3 mm Hg, P<0.0001; in white subjects: -13.6±3.1 versus 1.20±4.4, P<0.0001). In both racial groups, the good and poor responders did not differ significantly in mean age, previous treatment with antihypertensive medications, weight, body mass index, baseline diastolic BP, or baseline 24-hour urinary excretion of sodium. However, the poor responders to diuretic therapy had significantly greater mean 24-hour urinary excretion of aldosterone. Among black subjects but not white subjects, poor responders also had significantly greater mean duration of diagnosed hypertension, waist circumference, baseline systolic BP, plasma renin activity, serum aldosterone, and increase in 24-hour urinary excretion of sodium and lower mean serum potassium than the good responders (Table 1).
Table 1.
Characteristics of Responders and Nonresponders to Diuretic Therapy From the Genetic Epidemiology of Responses to Antihypertensives Study
| Black Subjects (N=194) |
White Subjects (N=195) |
|||||
|---|---|---|---|---|---|---|
| Characteristic | Good Response |
Poor Response |
Contrast P | Good Response |
Poor Response |
Contrast P |
| N | 97 | 97 | 98 | 97 | ||
| Women/men | 50/47 | 50/47 | 1.000 | 42/56 | 42/55 | 0.950 |
| Age, mean±SD, years | 47.1±6.1 | 47.7±6.5 | 0.523 | 48.4±7.9 | 48.7±6.8 | 0.774 |
| HTN duration, mean±SD, years | 5.87±5.61 | 10.64±8.38 | <0.0001 | 4.14±5.79 | 4.94±6.43 | 0.345 |
| HTN medication, mean±SD, n (%) | 85 (87.6) | 92 (94.9) | 0.076 | 87 (88.8) | 92 (94.9) | 0.123 |
| Weight, mean±SD, kg | 89.6±18.8 | 93.6±21.7 | 0.178 | 91.5±16.9 | 95.1±21.2 | 0.188 |
| BMI, mean±SD, kg/m-2 | 30.5±5.9 | 32.3±7.3 | 0.060 | 31.0±5.1 | 31.7±6.0 | 0.355 |
| Waist, mean±SD, cm | 94.5±11.6 | 99.2±16.6 | 0.026 | 103.2±12.8 | 104.1±16.6 | 0.679 |
| Systolic BP, mean±SD, mm Hg | 147.7±14.3 | 153.9±15.0 | 0.004 | 142.1±12.1 | 143.0±13.0 | 0.610 |
| Systolic BP response, mean±SD, mm Hg | -26.0±12.1 | -9.5±10.2 | <0.0001 | -17.8±11.0 | -3.9±10.9 | <0.0001 |
| Systolic BP response, mean±SD, adjusted | -26.6±9.6 | -8.3±9.6 | <0.0001 | -17.9±8.3 | -3.5±9.9 | <0.0001 |
| Diastolic BP, mean±SD, mm Hg | 97.0±5.4 | 97.3±5.6 | 0.759 | 95.6±5.2 | 95.6±6.1 | 0.996 |
| Diastolic BP response, mean±SD | -18.3±4.9 | -0.3±4.4 | <0.0001 | -13.7±4.2 | 1.1±5.3 | <0.0001 |
| Diastolic BP response, mean±SD, adjusted | -18.3±4.2 | -0.18±4.3 | <0.0001 | -13.6±3.1 | 1.20±4.4 | <0.0001 |
| Potassium, mean±SD, mmol/L-1 | 3.82±0.28 | 3.72±0.33 | 0.027 | 4.01±0.26 | 4.02±0.48 | 0.897 |
| Potassium response, mean±SD | -0.17±0.35 | -0.18±0.37 | 0.825 | -0.22±0.36 | -0.23±0.47 | 0.924 |
| Renin, mean±SD, ng · mL-1 · h-1 | 0.70±0.71 | 1.12±1.29 | 0.007 | 1.33±0.88 | 1.64±1.41 | 0.070 |
| Renin response, mean±SD | 1.60±2.02 | 1.71±3.00 | 0.778 | 1.14±1.77 | 1.24±2.87 | 0.766 |
| Aldosterone, mean±SD, ng/dL-1 | 15.8±8.6 | 19.1±10.7 | 0.017 | 14.5±6.8 | 15.4±10.2 | 0.482 |
| Aldosterone response, mean±SD | 4.8±7.5 | 4.2±9.1 | 0.602 | 4.6±9.8 | 2.7±9.2 | 0.181 |
| Urine aldosterone, mean±SD, μg · (24 h)-1 | 8.18±4.96 | 10.73±6.91 | 0.004 | 8.67±4.70 | 11.04±7.69 | 0.010 |
| Urine aldosterone response, mean±SD | 2.29±4.86 | 1.66±5.78 | 0.410 | 4.63±7.99 | 1.87±8.72 | 0.022 |
| Urine sodium, mean±SD, mmol · (24 h)-1 | 166.7±61.1 | 161.1±71.3 | 0.552 | 159.2±65.5 | 160.2±72.4 | 0.919 |
| Urine sodium response, mean±SD | -9.7±71.1 | 13.2±88.2 | 0.049 | -1.55±70.5 | 15.64±76.58 | 0.105 |
| Urine potassium, mean±SD, mmol · (24 h)-1 | 48.6±19.1 | 51.2±18.7 | 0.345 | 60.8±25.7 | 70.7±28.8 | 0.012 |
| Urine potassium response, mean±SD | 4.6±23.6 | 7.9±19.7 | 0.296 | 6.84±26.31 | 0.67±30.50 | 0.132 |
P value is significance of contrast between good and poor responders.
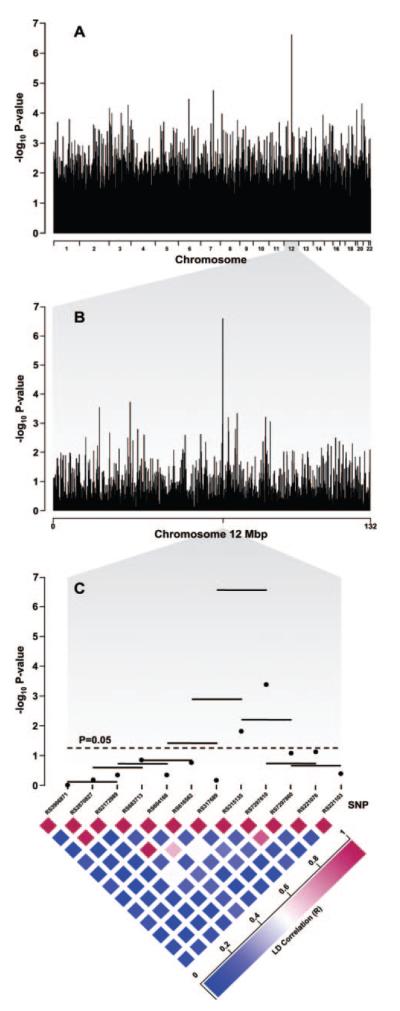
In black subjects, haplotype trend logistic regression across the 22 autosomes, using a 3-SNP haplotype sliding window, identified a region of chromosome 12q15, defined by haplotypes constructed from rs317689, rs315135, and rs7297610, that was most significantly associated with diastolic BP response category and yielded the smallest nominal P value genome wide (P=2.39×10-7; Figure 1). This P value was 74 times smaller than the next most significantly associated 3-SNP haplotype in black subjects (on chromosome 7q21.3: P=1.77×10-5) and 24 times smaller than the most significantly associated 3-SNP haplotype in white subjects (on chromosome 8p11.22: P=5.80×10-6; analyses not shown). After Bonferroni correction for the number of haplotype trend regression tests performed, only the chromosome 12q15 haplotypes remained significantly associated with adjusted diastolic BP response category in black subjects (ie, P=0.023, corrected for 94 924 haplotype trend regression tests in black subjects). The experiment-wise P value of the observed association, based on 1120 genome-wide permutation tests, was 0.040 (upper 95% confidence limit: 0.051). A score test of differences in haplotype frequencies19 between good and poor responders indicated that the ATC haplotype on chromosome 12q15 was significantly more frequent among black good responders (P=0.0002), whereas the ACT and the ATT haplotypes were significantly more frequent among black poor responders (P=0.0018 and P=0.0219, respectively; Table 2), suggesting that the third SNP, rs7297610 (T→C), may contribute the most to the observed association. This inference is also supported by the observation that rs7297610 had the most statistically significant single-site P value of the 3 SNPs (P=0.00036; Figure 1).
Figure 1.
Plots of P values from haplotype trend logistic regression analysis in the sample of 194 black subjects with essential hypertension, consisting of 97 good responders and 97 poor responders to hydrochlorothiazide. The -log10 P values are plotted as a function of location across the 22 autosomes (A, top), across chromosome 12 (B, middle), and across a region of chromosome 12q15 designated by reference sequence numbers of the SNPs genotyped on the Affymetrix 100K Human Mapping Array Set (C, bottom). In the bottom panel, separate horizontal lines depict the -log10 P value of each 3-SNP haplotype window above the component SNPs, and separate dots depict the -log10 P value of each SNP.
Table 2.
Estimated Frequencies of Haplotypes Constructed From Affymetrix 100K SNPs on Chromosome 12q15 Region (rs317689, rs315135, and rs7297610) Associated With Diastolic BP Response Category in Black Subjects
| Observed Haplotypes |
|||||
|---|---|---|---|---|---|
| Response Category | ACT | ATT | ACC | GTC | ATC |
| Overall | 5.6 | 23.2 | 6.0 | 12.7 | 52.5 |
| Poor responders | 8.7 | 28.1 | 6.8 | 13.4 | 43.0 |
| Good responders | 0.0 | 17.6 | 5.9 | 8.6 | 64.7 |
| P | 0.0018 | 0.0219 | 0.421 | 0.660 | 0.00017 |
Haplotype frequencies were estimated for the good and poor responders using Haplo.Stat software, and statistical significance of differences between groups was based on a score test.19
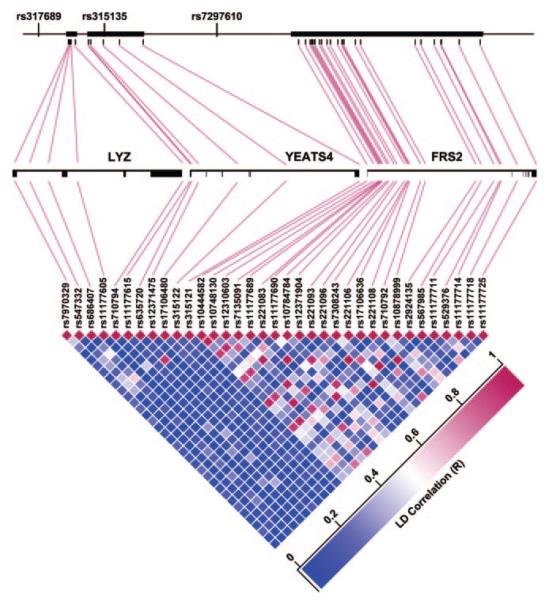
To localize or “fine map” the DNA sequence variation within genes in the chromosome 12q15 region that may account for association with diastolic BP response, we genotyped 35 SNPs selected to tag linkage disequilibrium blocks within the genes closest to the 3 Affymetrix SNPs (Figure 2), namely, LYZ, located downstream of the first SNP (rs317689); YEATS4, in which the second SNP (rs315135) is located; and FRS2, located downstream of the third SNP (rs7297610) (Figure 2). In black subjects, all 35 of the sites were polymorphic. Among the original 194 black good and poor responders, haplotype trend logistic regression analyses using a 3-SNP sliding window within each gene indicated that variation in YEATS4 was most significantly associated with diastolic BP response category (P=0.0124 for haplotype constructed with rs635720, rs12371475, and rs17106480). This was also supported by results of genecentric analyses of haplotypes constructed from all of the SNPs within in each gene, wherein variation in YEATS4 remained more significantly associated with diastolic BP response category (P=0.063) than variation in LYZ (P=0.108) or FRS2 (P>0.5; analyses not shown).
Figure 2.
Three genes in the chromosome 12q15 region: LYZ, YEATS4, and FRS2. The top portion depicts location and lengths of the genes, represented by heavy bars, in relationship to each other and to the 3 SNPs from the Affymetrix 100K Human Mapping Array Set, used to compose the 3-SNP haplotype significantly associated with diastolic blood pressure response category in black subjects (see Figure 1). The middle portion depicts the arrangement of introns and exons within each gene and the locations of 35 tag SNPs genotyped for fine mapping of the region. The bottom portion depicts pairwise linkage disequilibrium between the 35 tag SNPs.
Because the genecentric haplotype analyses described above implicated variation in LYZ and YEATS4 more strongly than in FRS2, further assessments of the genetic association between the chromosome 12q15 region and measures of diastolic BP response were limited to analyses of haplotypes constructed from the 11 tag SNPs in LYZ and YEATS4. In the statistically independent test data set derived from all 291 of the non-Hispanic black subjects, a 1—degree-of-freedom test of the 11-SNP haplotypes of LYZ+YEATS4, which provided optimal prediction of diastolic BP response category in the “training sample” of 194 black good and poor responders, yielded a 1-sided P value of 0.024 for association with the quantitative measure of residual variation in diastolic BP response (after subtracting off the group mean for the response category to which each subject belonged). In an independent data set consisting of all 294 of the non-Hispanic white subjects, a global F test of the 5-SNP haplotypes of LYZ yielded a P value of 0.039 for association with the quantitative measure of diastolic BP response.
Discussion
In contrast to candidate gene studies, which test a priori hypotheses that variation in particular genes influences a phenotype of interest, whole genome scans represent a fundamentally different, entirely “data-driven” approach to identifying novel pharmacogenetic loci and, as such, are unbiased by pre-existing knowledge regarding the location or identity of genes influencing drug response. We compared the good and poor responders to hydrochlorothiazide at >100 000 SNPs located across the 22 autosomes. Phenotypically distinct groups were sampled from opposite tertiles of the distribution of diastolic BP responses, because BP response is not measured without error,20 to reduce misclassification and thereby increase the power to detect genetic associations. Although the 100 000 SNP arrays represent only a small fraction of all of the polymorphisms described in the human genome, we assembled the measured SNPs into longer combinations of blocks of coinherited sequences, ie, haplotypes, constructed from successive 3-SNP windows, so that variation across larger regions spanning the 22 autosomes could be analyzed for associations with the categorical difference in BP response. In the primary genome scans, which were conducted in each race separately because of expected differences in haplotype frequencies, variation in only 1 region on chromosome 12q15 emerged to be significantly associated with BP response in black subjects after controlling for multiple testing. Follow-up haplotype analyses of 35 tag SNPs in the 3 genes in the region appeared to exclude FRS2 and to favor YEATS4 over LYZ as positional candidate genes to influence BP response to hydrochlorothiazide.
Because many candidate gene polymorphisms initially reported to be associated with common, complex disorders, such as hypertension, were not confirmed in subsequent studies, such reports may be “false-positives” or a reflection of small, context-dependent genetic effects.21,22 Replication of a positive genetic association in another sample from the same population protects against false-positives that may elude statistical controls, whereas validation in a sample from a different population supports generalizability of the observed association, ie, it is not unique to the context of the original study. In our fine mapping analyses of genes in the chromosome 12q15 region, variation in YEATS4 appeared to be most strongly associated with response category in the primary sample of 194 black good and poor responders. However, consideration of variation in both LYZ and YEATS4 was required to provide corroborating evidence of an association between variation in genes from this region with a statistically independent measure of diastolic BP response derived for the parent sample of all 291 of the black subjects. Moreover, consideration of haplotype variation limited to LYZ was required for validation of an association in an independent data set derived from all 294 of the white subjects. In lieu of a replicate sample and because these additional analyses suggest that variation in YEATS4 alone is unlikely to account for the observed associations with BP response, additional confirmation of the associations in other independent samples is indicated before attempting to identify functional sites within ≥1 gene in the region.
Although genome scans have an undisputed advantage over candidate gene studies in providing an unbiased approach to identify novel genes influencing a phenotype of interest, interpretation of what appear to be novel “truepositive” discoveries based on statistical evidence can be more challenging than when variation in a particular gene or protein product is already known or suspected to influence the phenotype. For the positional candidate genes in the chromosome 12q15 region, we addressed this challenge by applying a modification of the Gene Set Enrichment Analysis method23 (please see the data supplement available online at http://hyper.ahajournals.org), which allowed us to leverage additional information from the 100 000 SNP arrays and take advantage of previously determined sets of genes known to be functionally or otherwise biologically interrelated. This approach was used to score 29 “gene loci sets” extracted from the Molecular Signature Database provided with the Gene Set Enrichment Analysis software, consisting of SNPs in genes related to LYZ, YEATS4, or FRS2.23 Of the 29 sets evaluated, only the “aging kidney up” set appeared to be associated with BP response to hydrochlorothiazide in white subjects. The aging kidney up gene set was so named because increasing age was associated with upregulation of the gene expression in normal human kidney tissue.24 This finding is intriguing insofar as the target of hydrochlorothiazide (ie, the sodiumchloride cotransporter) is located in the kidney (ie, luminal membrane of distal convoluted tubular cells), and greater BP response to thiazide diuretics has been associated with older age.6 However, the aging kidney up gene set was considered for this analysis because it contains FRS2, the gene from the chromosome 12q15 region for which evidence of association with BP response was weakest, and evidence of association of the aging kidney up genes, in aggregate, with BP response, was not observed in black subjects, the group in which evidence of the chromosome 12q15 association with BP response was strongest. Results of this exploratory analysis may be consistent with a multilocus effect of aging kidney up genes on drug response in white subjects but a single locus effect in black subjects, limited to the chromosome 12q15 region. Additional investigation is warranted to further support this hypothesis and to elucidate the biological mechanism that may underlie the observed associations with BP response to hydrochlorothiazide.
The present study has several limitations. First, heritability of antihypertensive drug responses has not been established in biometrical analyses of family based samples and may only be modest in magnitude, similar to that of BP level (ie, ≅0.3). Moreover, the genetic contribution to antihypertensive response to the thiazide diuretic may be polygenic (ie, many genes with small effects), also similar to BP level. Reliable detection of such a small genetic effect may require larger sample sizes. Second, the convenience advantage of office measurements of BP, as performed in the present study, may be counterbalanced by greater error variability than ambulatory or home recordings. Third, the hydrochlorothiazide dosage was based on clinical practice guidelines and may not have been sufficient to overcome pharmacokinetic differences contributing to interindividual differences in drug response. Fourth, the follow-up genotyping of the chromosome 12q15 region was limited to tag SNPs within LYZ, YEATS4, and FRS2 and did not consider possible contributions of variation within intergenic regions.
Perspectives
Initial pharmacogenetic studies have associated several candidate gene polymorphisms with BP responses to a variety of antihypertensive drugs; however, none of the reported associations has yet been confirmed in subsequent studies. Because the phenotypic effects of reported pharmacogenetic associations appear to be either too small or too rare to be useful in routine clinical practice, current guidelines for the treatment of hypertension do not recommend genetic measurements. Results of this study provide an initial demonstration of the challenges and promises of genome-wide association analyses to localize novel genes influencing BP response to commonly prescribed antihypertensive medications.
Supplementary Material
Acknowledgments
We are thankful for the technical assistance of Zhiying Wang, Meagan Grove, Jodie Van De Rostyne, Jeremy Palbicki, Robert Tarrell, and Prabin Thapa.
Sources of Funding
This work was supported by HL 74735, HL 53335, and the Mayo Foundation.
Footnotes
Disclosures None.
References
- 1.Meissner I, Whisnant JP, Sheps SG, Schwartz GL, O’Fallon WM, Covalt JL, Sicks JD, Bailey KR, Wiebers DO. Detection and control of high blood pressure in the community: do we need a wake-up call? Hypertension. 1999;34:466–471. doi: 10.1161/01.hyp.34.3.466. [DOI] [PubMed] [Google Scholar]
- 2.Turner ST, Schwartz GL, Chapman AB, Hall WD, Boerwinkle E. Antihypertensive pharmacogenetics: getting the right drug into the right patient. J Hypertens. 2001;19:1–11. doi: 10.1097/00004872-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Laragh JH, Lamport B, Sealey J, Alderman MH. Diagnosis ex juvantibus. Individual response patterns to drugs reveal hypertension mechanisms and simplify treatment. Hypertension. 1988;12:223–226. doi: 10.1161/01.hyp.12.3.223. [DOI] [PubMed] [Google Scholar]
- 4.Turner ST, Schwartz GL, Boerwinkle E. Personalized medicine for high blood pressure. Hypertension. 2007;50:1–5. doi: 10.1161/HYPERTENSIONAHA.107.087049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr, Jones DW, Materson BJ, Oparil S, Wright JT, Jr, Roccella EJ, the National High Blood Pressure Education Program Coordinating Committee Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension. 2003;42:1206–1252. doi: 10.1161/01.HYP.0000107251.49515.c2. [DOI] [PubMed] [Google Scholar]
- 6.Materson BJ, Reda DJ, Cushman WC, Massie BM, Freis ED, Kochar MS, Hamburger RJ, Fye C, Lakshman R, Gottdiener J, Ramirez EA, Henderson WG. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. N Engl J Med. 1993;328:914–921. doi: 10.1056/NEJM199304013281303. [DOI] [PubMed] [Google Scholar]
- 7.Preston RA, Materson BJ, Reda DJ, Williams DW, Hamburger RJ, Cushman WC, Anderson RJ. Age-race subgroup compared with renin profile as predictors of blood pressure response to antihypertensive therapy. Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. JAMA. 1998;280:1168–1172. doi: 10.1001/jama.280.13.1168. [DOI] [PubMed] [Google Scholar]
- 8.Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. WNK1 kinase polymorphism and blood pressure response to a thiazide diuretic. Hypertension. 2005;46:758–765. doi: 10.1161/01.HYP.0000186240.81996.57. [DOI] [PubMed] [Google Scholar]
- 9.Cusi D, Barlassina C, Azzani T, Casari G, Citterio L, Devoto M, Glorioso N, Lanzani C, Manunta P, Righetti M, Rivera R, Stella P, Troffa C, Zagato L, Bianchi G. Polymorphisms of alpha-adducin and salt sensitivity in patients with essential hypertension. Lancet. 1997;349:1353–1357. doi: 10.1016/S0140-6736(97)01029-5. [DOI] [PubMed] [Google Scholar]
- 10.Chapman AB, Schwartz GL, Boerwinkle E, Turner ST. Predictors of antihypertensive response to a standard dose of hydrochlorothiazide for essential hypertension. Kidney Int. 2002;61:1047–1055. doi: 10.1046/j.1523-1755.2002.00200.x. [DOI] [PubMed] [Google Scholar]
- 11.Turner ST, Chapman AB, Schwartz GL, Boerwinkle E. Effects of endothelial nitric oxide synthase, alpha-adducin, and other candidate gene polymorphisms on blood pressure response to hydrochlorothiazide. Am J Hypertens. 2003;16:834–839. doi: 10.1016/s0895-7061(03)01011-2. [DOI] [PubMed] [Google Scholar]
- 12.Cutler DJ, Zwick ME, Carrasquillo MM, Yohn CT, Tobin KP, Kashuk C, Mathews DJ, Shah NA, Eichler EE, Warrington JA, Chakravarti A. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11:1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 16.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 17.Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- 18.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 19.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkielman JD, Schwartz GL, Chapman AB, Boerwinkle E, Turner ST. Reproducibility of blood pressure response to hydrochlorothiazide. J Clin Hypertens (Greenwich) 2002;4:408–412. doi: 10.1111/j.1524-6175.2002.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 22.Manly KF. Reliability of statistical associations between genes and disease. Immunogenetics. 2005;57:549–558. doi: 10.1007/s00251-005-0025-x. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, Myers BD, Brooks JD, Davis RW, Higgins J, Owen AB, Kim SK. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.