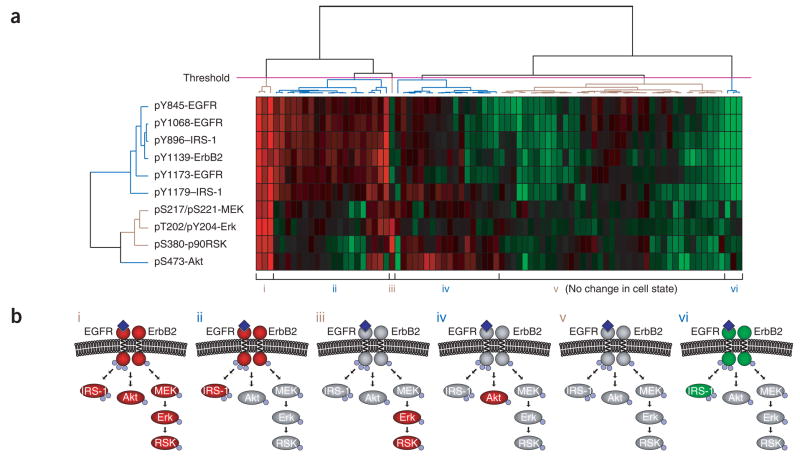
Figure 4.
Clustering of data from a state-based screen. (a) Heat map obtained by hierarchical clustering of relative inhibition values for each small-molecule–antigen pair. Positive inhibition values, indicating a decrease in phosphorylation, range from 0 (black; no inhibition) to 1 (red; 100% inhibition). Negative inhibition values, indicating an increase in phosphorylation, range from 0 (black; no inhibition) to −1 (green; twofold increase in phosphorylation). (b) Simplified view of the six network states defined by the threshold (magenta line) shown in a. Red ovals represent proteins with decreased phosphorylation; green ovals represent proteins with increased phosphorylation; gray ovals represent proteins whose phosphorylation was unaffected. Small blue spheres represent sites of phosphorylation. Dark blue diamonds represent EGF bound to the extracellular domain of EGFR.