Abstract
The exocyst is a eukaryotic tethering complex necessary for the fusion of exocytic vesicles with the plasma membrane. Its in vivo function is tightly regulated by interactions with multiple small GTPases. Exo70, one of the eight subunits of the exocyst, is important for the localization of the exocyst to the plasma membrane. It interacts with TC10 and Rho3 GTPases in mammals and yeast, respectively, and has recently been shown to bind to the actin-polymerization complex Arp2/3. Here we present the crystal structure of Mus musculus Exo70 at 2.25Å resolution. Exo70 is composed of α-helices in a series of right-handed helix-turn-helix motifs organized into a long rod of length 170Å and width 35Å. Although the α-helical organization of this molecule is similar to that in Saccharomyces cerevisiae Exo70, major structural differences are observed on the surface of the molecule, at the domain boundaries, and in various loop structures. In particular, the C-terminal domain of M. musculus Exo70 adopts a new orientation relative to the N-terminal half not seen in S. cerevisiae Exo70 structures. Given the low level of sequence conservation within Exo70, this structure provides new insights into our understanding of many species-specific functions of the exocyst.
Keywords: Crystallography, Membrane Trafficking, Exocytosis, Exocyst, Exo70
INTRODUCTION
Intracellular vesicle delivery depends upon a series of protein complexes working in tandem to transport, tether, and fuse vesicles to specific target membranes. Tethering complexes are thought to mediate the SNARE-dependent fusion of these vesicles to the target membrane at specific sites. In exocytosis, exocytic vesicles are transported from the Golgi apparatus along the cytoskeleton to specific fusion sites on the plasma membrane (PM) by the unconventional myosin Myo2 in yeast1,2 and others in mammals. The exocyst is a multi-subunit tethering complex involved in exocytosis and receptor recycling3–6. It has been identified in most eukaryotes7 and is composed of eight protein subunits: Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo843,8–10. The exocyst is the first contact between the vesicle and the PM in exocytosis and is known to regulate exocytosis through interactions with multiple small GTPases in a GTP-dependent manner, including Rho111, Rho312, Cdc4213, and Sec414 in yeasts; and RalA15–18, TC1019, Arf65, and Rab1120 in mammals. Disruption of the exocyst function does not affect the ability of exocytic vesicles to reach the PM, but it does affect the ability of these vesicles to fuse with the PM21,22.
Exo70 is the 70kDa subunit of the exocyst and its primary sequence is only weakly conserved among Exo70 orthologs (16% identity and 35% similarity between Mus musculus and Saccharomyces cerevisiae). Within the exocyst, Exo70 is known to interact with Sec623,24, Sec823,25, and Sec1023,25. It also interacts with Sec15 and Exo84 in mammals24 and Sec5 in yeast14. More recently, Arpc1, a subunit of the Arp2/3 complex, has been shown to interact with Exo70, and this interaction is important for actin network reorganization at the PM in both yeast and mammals26. Exo70 also mediates interactions between the exocyst and small GTPases, which appears to play a role in the regulation and assembly of the exocyst. Interaction with activated TC10 regulates the presence of exocytic sites at lipid rafts in insulin-stimulated glucose transport in mammals19,27. In yeast, interaction with activated Rho3 controls the fusion of exocytic vesicles with the PM12,28 and may also be important in demarcation of exocytic sites29. Despite some overlapping functions, however, interaction with these small GTPases occurs at two separate sites on Exo7019,23, suggesting that the functional organization of Exo70 may not be entirely conserved. While the recently reported structures of S. cerevisiae Exo70 (ScExo70)23,30 have been used as a general model to understand the structure and function relationship of all Exo70 proteins, it is essential to obtain high-resolution structural information of mammalian Exo70 in order to address questions specific to mammalian exocytosis.
Here we report the crystal structure of M. musculus Exo70 (MmExo70) at 2.25Å resolution. Comparison to the structure of ScExo70 reveals that while the overall fold of the protein is conserved, there is significant structural reorganization within the molecule that results in the reorientation of the C-terminal domain relative to the rest of the molecule. In addition, loops connecting secondary structural elements show significant differences in length and conformation. This, combined with a general lack of primary sequence conservation, results in the alteration of the molecule’s surface properties. The observed structural similarity and differences are consistent with the function of Exo70 as an exocyst subunit that interacts with different sets of effector molecules in different organisms. The structure of MmExo70 is the first structure of a mammalian exocyst subunit and makes Exo70 the first exocyst subunit with structural information available in more than one organism.
RESULTS AND DISCUSSION
Structure determination
To obtain diffraction-quality crystals of MmExo70, purified full-length protein was subjected to limited proteolysis by subtilisin, which identified a major fragment containing a deletion of the N-terminal 84 residues of the molecule. This fragment (residues 85–653) was crystallized in the P3221 space group with unit cell dimensions of a = b = 61.5Å, c = 294.7Å and one molecule in the asymmetric unit. The structure was determined by the multi-wavelength anomalous diffraction (MAD) method using crystals grown from L-selenomethionine-substituted protein. The final model was refined to a resolution of 2.25Å with an R-factor of 23.5% and an Rfree of 28.5% (Table 1), which contains residues 85–179, 188–241, 275–446, 455–652, and 170 water molecules. There is no clear electron density observable for residues 180–187, 242–274, 447–454, and 651–653. These residues are presumably disordered. Of all residues in the structure, 92.8% are found in the most favored regions of the Ramachandran plot31, while the rest are found in either the additionally allowed (6.5%) or the generously allowed (0.6%) regions.
Table 1.
Crystallographic data statistics for MmExo70
| MmExo70 Native | MmExo70 L-Selenomethionine substituted | |||
|---|---|---|---|---|
| Data collection | ||||
| Space group | P3221 | P3221 | ||
| Cell dimensions [a, b, c (Å)] | 61.52, 61.52, 294.73 | 61.64, 61.64, 294.68 | ||
| Resolution (Å) | 50.0–2.25 (2.33–2.25) | 50.0–2.5 (2.59–2.50) | ||
| Peak | Inflection | Remote | ||
| Wavelength (Å) | 0.97926 | 0.97926 | 0.97942 | 0.95660 |
| Completeness (%) | 93.7 (77.7) | 98.6 (89.2) | 97.1 (77.5) | 93.4 (59.2) |
| Redundancy | 4.2 (3.1) | 6.2 (4.6) | 5.7 (3.0) | 5.4 (2.3) |
| I/σI | 24.6 (3.0) | 34.0 (4.7) | 32.0 (2.8) | 29.8 (1.8) |
| Rmerge(%) | 5.4 (41.6) | 7.9 (29.7) | 6.9 (36.5) | 6.9 (42.3) |
| Refinement | ||||
| Resolution (Å) | 50.0–2.25 | |||
| No. reflections in working set | 27369 | |||
| No. reflections in test set | 1435 | |||
| Rwork/Rfree (%) | 23.5/28.5 | |||
| No. of atoms | ||||
| Protein | 4099 | |||
| Water | 170 | |||
| Average B-factors | ||||
| Protein | 54.4 | |||
| Water | 54.8 | |||
| Rms deviations from ideality | ||||
| Bond lengths (Å) | 0.006 | |||
| Bond angles (deg.) | 1.1 | |||
| Ramachandran plot | ||||
| % in most favored regions | 92.8 | |||
| % in addl. allowed regions | 6.5 | |||
| % in gen. allowed regions | 0.6 | |||
Highest resolution shell shown in parentheses.
Structural organization of MmExo70
MmExo70 is composed of nineteen α-helices (H1-H19) connected by loops of varying lengths (L1–2-L18–19; numbers identify adjoining α-helices) organized into three distinct domains to form a 170 Å-long, 35 Å-wide rod-shaped molecule (Figure 1a, 2). The N domain is composed of residues 85–393, including H1-H9 and the first half of H10. These α-helices are organized into a series of right-handed helix-turn-helix motifs with a slight super-helical twist that extends nearly 100Å. H4 is the shortest helix in this domain and is an exception to the helix-turn-helix motif as it is perpendicular to the other α-helices. Most of the loops in this domain are short (2–5 residues) except for L4–5, a partially disordered loop of sixteen-residues, and L6–7, which contains up to 33 disordered residues and is predicted to be largely unstructured32. The M domain is composed of residues 394–538, including the second half of H10, H11-H15, and the first half of H16. These helices are organized into a 55Å-long five-helix bundle that maintains the right-handed helix-turn-helix motif. The long axis of this domain is canted about 40° from the long axis of the full molecule. H12 and H15 are two short helices that are not part of the α-helical bundle structure, as they both closely follow the preceding helix with an approximate 90° turn. The loops of the M domain are short except for L10–11 and L12–13, which are 15 and 10 residues long, respectively, with most of L12–13 being disordered. The C domain is composed of residues 539–650, including the second half of H16 and H17–H19. These helices are organized into a 55Å-long four-helix bundle that continues the right-handed helix-turn-helix motif. The boundary between the M and C domains is defined by a kink in H16 that orients this domain along the long axis of the molecule.
Figure 1. The Structure of Exo70.
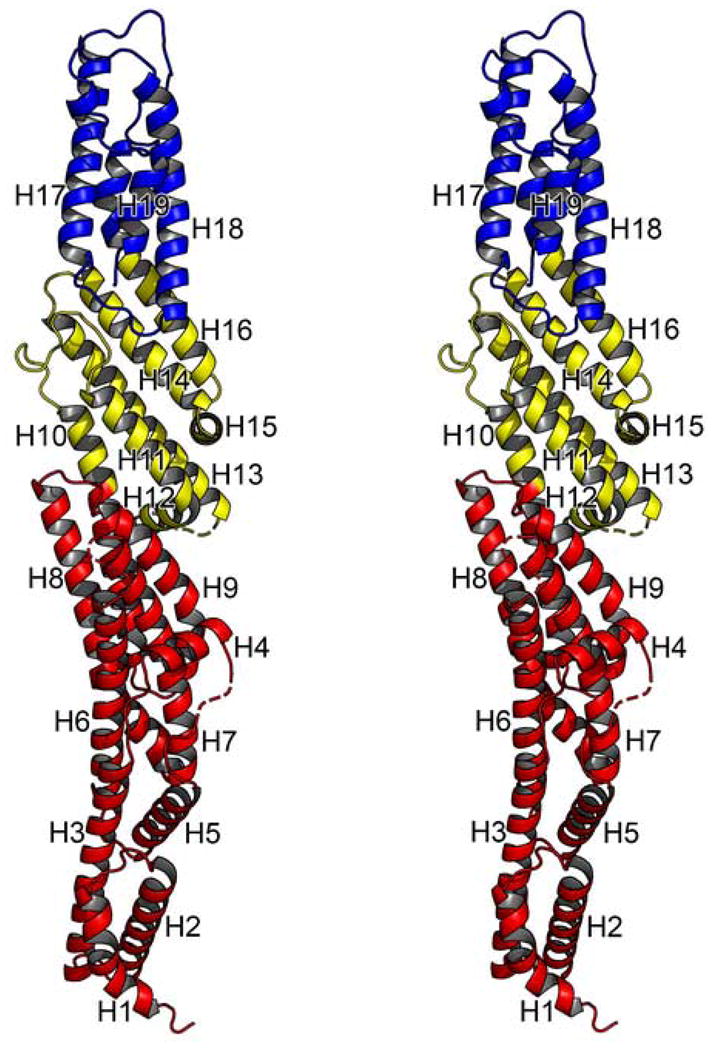
Stereo cartoon diagram of MmExo70 is shown. Residues of the N domain are shown in red, M domain in yellow, and C domain in blue. α-helices are drawn as coils and labeled from H1 to H19; turns and loops, solid tubes; unobserved residues, dashed lines (no β strands are observed in the structure).
Figure 2. Structural Alignment of Exo70 Proteins.
Structural alignment of Exo70 in M. musculus, H. sapiens, D. melanogaster, S. pombe, and S. cerevisiae. Invariant residues are shaded purple. Similar but not identical residues are shaded pink. Secondary structural elements are indicated above the sequence block for MmExo70 and below for ScExo70: α-helices, rectangles; other elements, solid lines; structurally unobserved residues, dashed lines. For MmExo70, N domain, red; M domain, yellow; and C domain, blue (same as Figure 1). For ScExo70, N domain, brown; M domain, green; and C domain, cyan. For clarity, sequences lacking homology or unobserved in the structures have been replaced with bracketed numbers indicating the number of residues omitted.
The interface between the N and M domains, consisting of residues from H8-H9 and H10-H12, appears as a thin “neck” with a buried surface area of about 740Å2 (Figure 3a). It is composed of two small hydrophobic patches that each contains a cluster of mostly aromatic residues and a few surrounding water-mediated hydrogen bonds. The first hydrophobic patch includes Phe351 (H9) packing against Phe434 (H12). The second hydrophobic patch, separated by 6Å from the first, consists of Phe344 (L8–9) packing against Phe397 (H10) and Phe427 (H11). The hydrogen bonds involve the main chain carbonyl group of Ile339 (H8) and the side chain of His342 (L8–9) interacting with the side chain of Asn400 (H10), the side chain of Arg355 (H9) interacting with the main chain carbonyl group of Asp433 (H11) and the side chain of Glu436 (H12), and the side chain of Glu387 (H10) interacting with the side chain of Gln445 (H12).
Figure 3. Domain boundaries of MmExo70.
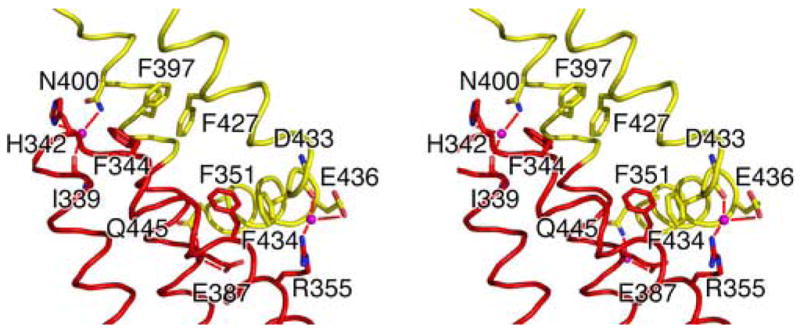
Stereo cartoon diagrams showing interactions between the N and M domains (A) and M and C domains (B) of MmExo70. The side chains or main chains of residues involved in domain-domain hydrogen bonds are shown as stick models with oxygen and nitrogen atoms colored red and blue, respectively. The rest of the protein structure is shown as a ribbon drawing and is colored by domain as in Figure 1. Solvent molecules involved in hydrogen bonds are shown as magenta spheres. Hydrogen bonds are depicted as red lines. Aromatic residues involved in the hydrophobic patches are also included in (A). Helices other than H8-H12 are omitted in (A) and H19 is omitted in (B) for clarity.
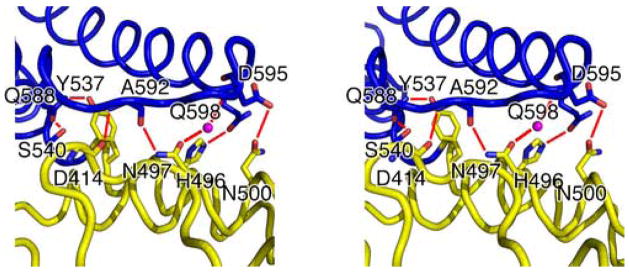
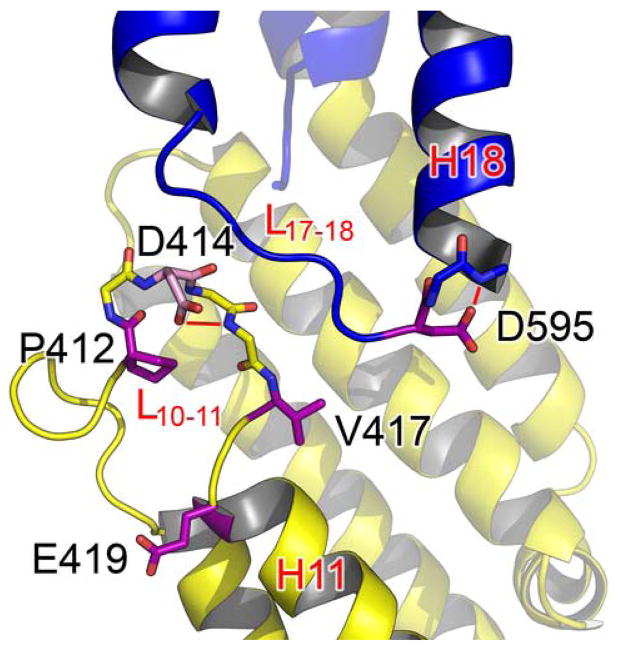
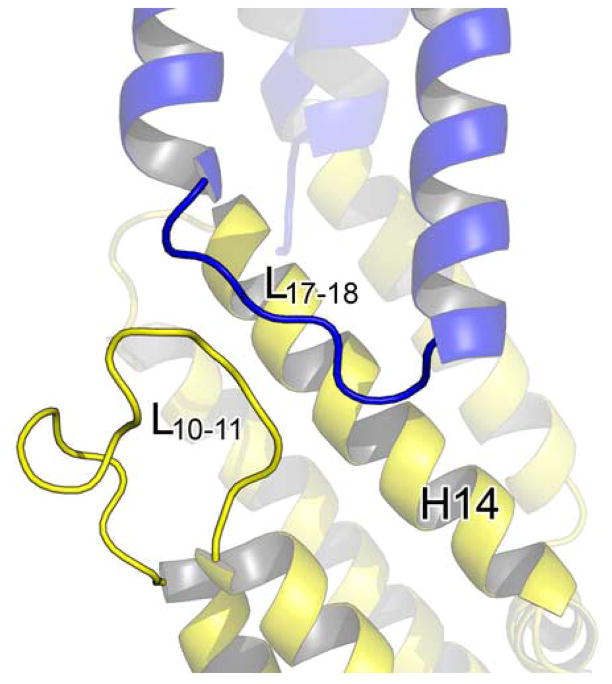
The interface between the M and C domains primarily involves L10–11, H14, and H16 packing against H17-H18 and buries about 755Å2 of surface area (Figure 3b). Many hydrophobic residues on H14 and H16 interact with residues on H17-H18, composing the core of this interface. In addition, a number of hydrogen bond interactions also stabilize the domain interface. These include main chain interaction between the carbonyl group of Asp414 (L10–11) and the amide group of Ala592 (L17–18), side chain interaction between His496 (H14) and Gln598 (H18), side chain interaction between Asn500 (H14) and Asp595 (L17–18), dual side chain interactions between Gln588 (H17) and Tyr537/Ser540 (H16), and water mediated dual hydrogen bonds between the side chain of Asp497 (H14) and the main chain carbonyl group of Ala592/the main chain amide group of Asp595 (L17–18). Interestingly, L10–11 and L17–18 are parallel to one another for a length of 11Å but interact only through the one main chain hydrogen bond mentioned above.
Conserved residues of Exo70
The low level of primary sequence conservation among Exo70 orthologs makes previous sequence alignment, and therefore identification of conserved residues, unreliable. The structure of MmExo70 enables, for the first time, structure-based sequence alignment to examine the role of conserved residues in Exo70 (Figure 2). For structural comparison with ScExo70 throughout this study, a crystal structure of ScExo70 determined in our own lab was used (see supplementary information). Briefly, this ScExo70 structure was determined to a resolution of 2.1 Å, which is similar to the published P212121 form23 and much higher than the published C2 form30. The overall structure is nearly identical to that of the P212121 form, with a root-mean square distance (rmsd) of 0.4 Å for 532 Cα atoms. Our structure, however, is the most complete Sc Exo70 structure available, including seventeen additional residues not present in the P212121 form and twenty-six additional residues not present in the C2 form.
Structural alignment reveals two regions with clusters of highly conserved residues. The first region is concentrated on L10–11 and H11 (Figure 4a). Pro412 is located at the start of a tight turn within L10–11 and is likely to be important for this structural feature. Asp414 is located at the end of the turn initiated by Pro412 and its side chain forms a hydrogen bond with the main chain amide of Thr416, possibly stabilizing this turn. As mentioned earlier, the main chain carbonyl group of Asp414 forms a hydrogen bond with the amide group of Ala592 on L17–18. This interaction is possibly stabilized by the side chain interactions of Asp414. Asp595 is located at the end of L17–18 and forms a hydrogen bond with the amide group of Glu597 on H18, possibly stabilizing the conformation of this loop. All of these interactions, except the main chain hydrogen bond between Asp414 and Ala592, appear to be conserved in the ScExo70 structure, suggesting that they are structurally important in supporting this part of the structure. Glu419 is the first residue of H11 and is completely exposed to solvent. The reason for its conservation is not clear but it could be involved in a conserved interaction. It is 12Å away from Asp414 and more than 22Å away from Asp595, the closest conserved surface-exposed charged residues. Finally, Thr421 on H11 forms a hydrogen bond with Asn498 on H14, which acts as a pivot point to allow rearrangement within the M domain (see below).
Figure 4. Conserved surface residues of MmExo70.
In both figures, completely conserved residues are colored purple, similar residues are colored pink, and the rest of the molecule is colored by domain as in Figure 1. (A) A cartoon diagram of the cluster of conserved residues found on the surface of the M domain of MmExo70. Main chain atoms are shown for P412-V417 and D595-E597 and side chains are shown for the conserved residues in stick models. Oxygen and nitrogen atoms are colored red and blue, respectively. Hydrogen bonds are depicted as red lines. Relevant α-helices and loops are labeled in red. The view is the same as in Figure 1. (B) Molecular surface diagram of the cluster of conserved residues found on the surface of the C domain of MmExo70. This view is rotated 45° about the long axis from Figure 1.
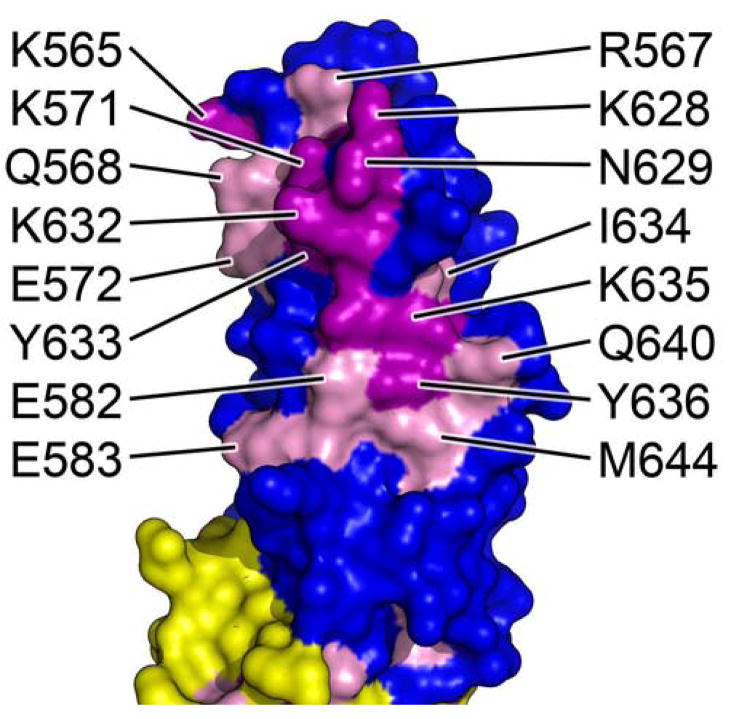
The second conserved region is a large (almost 400Å2), mostly hydrophilic surface patch found on H17, H19, and L18–19 (Figure 4b). Residues participating in this patch include Lys565, Arg567, Gln568, Lys571, Glu572, Glu582, Glu583 (H17), Lys628, Asn629, Lys632, Tyr633, Ile634, Lys635, Tyr636 (L18–19), Gln640, and Met644 (H19). Completely conserved residues appear mostly surrounded by similarly conserved residues. At least a portion of this region has been recently shown to interact with Arpc1 of the Arp2/3 complex. Mutation of Lys571 and Glu572, or Lys628, Asn629, and Pro630, abolishes the ability of Exo70 to interact with Arpc126. Presumably, all conserved residues on the surface of the C domain either directly interact with Arpc1 or stabilize the local structure. The hydrophobic core of the four-helix bundle is also well conserved and highly aromatic, contributing to the stability of this domain.
Structural Comparison to ScExo70
MmExo70 has an overall fold similar to ScExo7023,30 (Figure 5a). Each of the nineteen α-helices of MmExo70 corresponds closely to one of the nineteen α-helices of ScExo7023, although the structural differences are significant enough that attempts to determine the MmExo70 structure by molecular replacement using ScExo70 as a search model were not successful. Both molecules take the shape of a rod with similar dimensions23,30. The rod of ScExo70 is somewhat more curved and twisted than MmExo70. This curvature appears to originate primarily from the packing of helices H3-H5 against H6-H7, adding to the super-helical twist of the N domain, and from the packing of H11 and H13 against H14-H16 in the M domain, resulting in the twisting of the C domain away from the long axis of the molecule.
Figure 5. Differences in M domain packing affect M and C domain interactions.
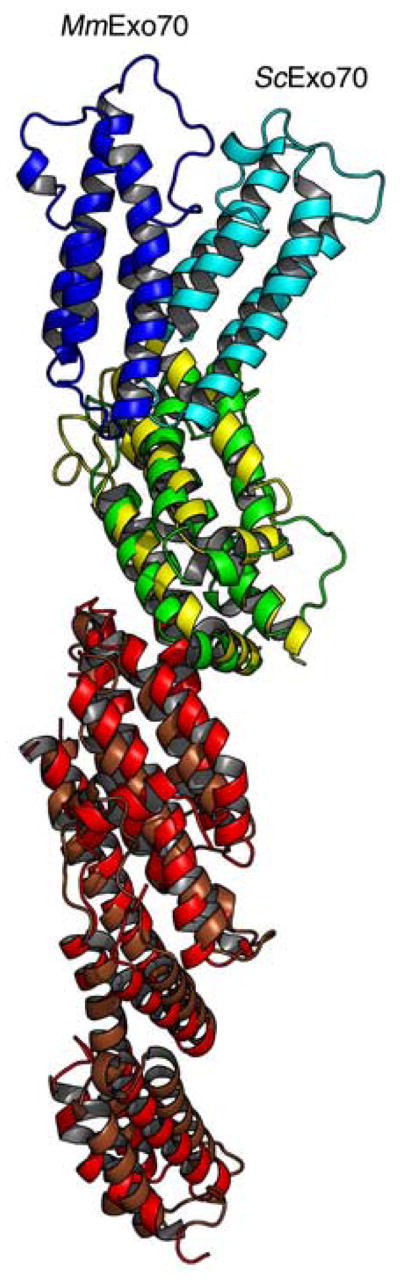
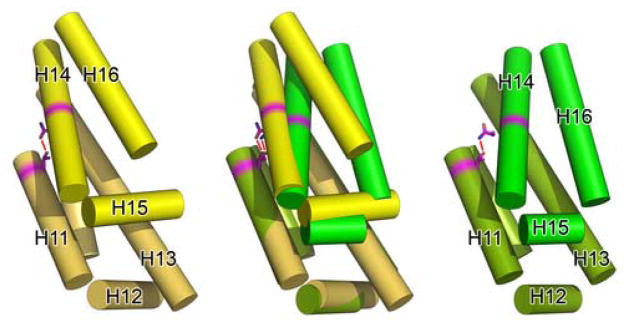
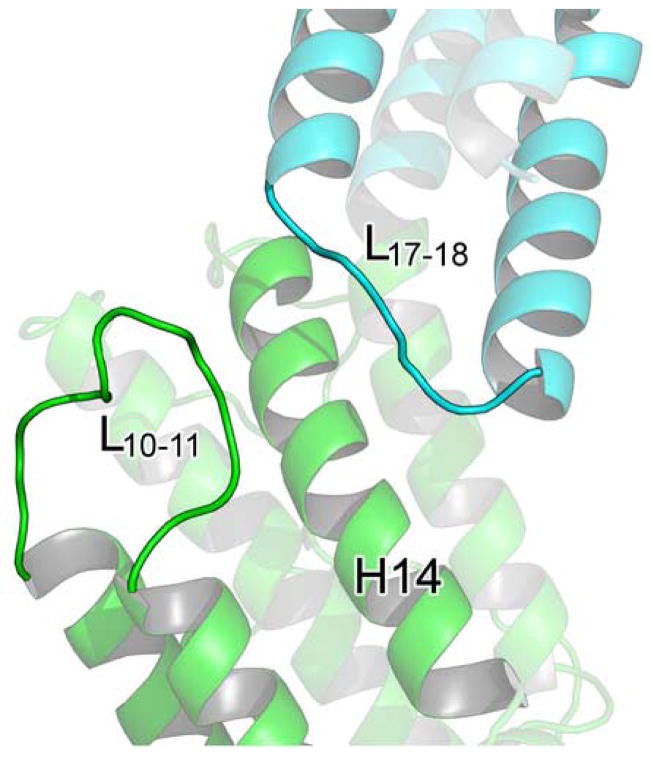
(A) A cartoon diagram showing the alignment of MmExo70 with ScExo70 based on alignment of the N domain. Residues are colored by domain as in Figure 2. (B) Schematic diagrams of the M domains of Exo70 with helices shown as cylinders and all loops omitted. The left panel shows MmExo70 (yellow) and the right panel shows ScExo70 (green). H11-H16 are labeled. The middle panel shows the alignment of MmExo70 with ScExo70 based on H10-H13. The completely conserved hydrogen bond between MmExo70 Thr421/ScExo70 Thr372 and MmExo70 Asn498/ScExo70 Asn479 is shown as a red line. Note the differences in α-helical packing between the two molecules. (C) A cartoon diagram showing the positions of L10–11 and L17–18 in MmExo70. Relevant α-helices and loops are labeled. (D) A cartoon diagram showing the positions of L10–11 and L17–18 in ScExo70. Relevant α-helices and loops are labeled.
The most notable difference between the two structures lies in the relative orientation of the C-terminal portion of the molecule to the N-terminal portion (Figure 5a). This originates from changes in the helical packing within the M domain. In support of this, the rmsd of the full MmExo70 and ScExo70 structures is 7.0Å for 481 Cα atoms, while the rmsd of H1-H13 is 3.6Å for 327 Cα atoms and the rmsd of H14-H19 is 2.9Å for 157 Cα atoms. Structural alignment of MmExo70 with ScExo70 based on H10-H13 reveals that H14-H16 adopts a conformation in which H14 and H16 are rotated 18.9° and 24.7°, respectively, between structures (Figure 5b). This alignment also reveals that the conformational change between the two halves of the molecule appears to pivot about Asn498 (H14), which forms a hydrogen bond with Thr421 (H11). Both of these residues are completely conserved and appear in a similar location in ScExo70, suggesting that this interaction is important to the internal packing of the M domain.
We argue that this conformational difference in the M domain between MmExo70 and ScExo70 is not a reflection of structural flexibility within the M domain. Rather, it suggests inherent structural differences between the two molecules. The thermal factors in this region are comparable to the rest of the molecule. Most importantly, the same conformation is observed in this region in all six independently determined ScExo70 structures23,30 (The C2 ScExo70 crystal form contains four molecules in the asymmetric unit). Furthermore, the MmExo70 conformation is stabilized by the previously mentioned interactions between M domain L10–11 and C domain L17–18 (Figure 5c). This interaction represents the major structural difference in an otherwise well-conserved region. Interestingly, the parallel organization of these loops is also observed in ScExo70 structures, although residues of these loops are separated for more than 4Å (Figure 5d). In comparison, the ScExo70 conformation is stabilized by interactions of L12–13 and L13–14 with H10, H13, and H16. The length of L13–14 may contribute to the observed conformational differences as well. The tight turn of the three- residue L13–14 in MmExo70 puts spatial constraints on the location of H14 while the eighteen-residue ScExo70 L13–14 grants more freedom to H14. Our structure-based sequence alignment reveals that Schizosaccharomyces pombe and higher eukaryotes have a nearly uniformly short L13–14 (Figure 2), suggesting that these organisms may share the same M domain packing and C domain orientation as observed in MmExo70.
Conformational differences are also observed at the interface between N and M domains. Major structural flexibility has been described for this interface in ScExo7030, which contains a similarly thin “neck” with almost 30% less buried surface area (580Å2 compared to 740Å2 in MmExo70)30 and is composed of only one hydrophobic patch and no hydrogen bonds. Phe306 (H9), Tyr385 (L11–12), Phe303 (H9), Phe345 (H10), and Phe382 (H11) in ScExo70 are structurally equivalent to Phe351 (H9), Phe434 (H12), Phe344 (H9), Phe397 (H10), and Phe427 (H11) in MmExo70 (Figure 3a). In addition, ScExo70 H10 is kinked at the domain boundary whereas no apparent kink is seen in MmExo70 H10, although this does not appear to affect the spatial relationship between the N and M domains or the orientation of the M domain. Currently, there is no evidence to suggest that a similar degree of flexibility also exists at the same domain in MmExo70, and the increased surface area and number of interactions would argue that it is more stable than in ScExo70.
Differences in surface electrostatic potential
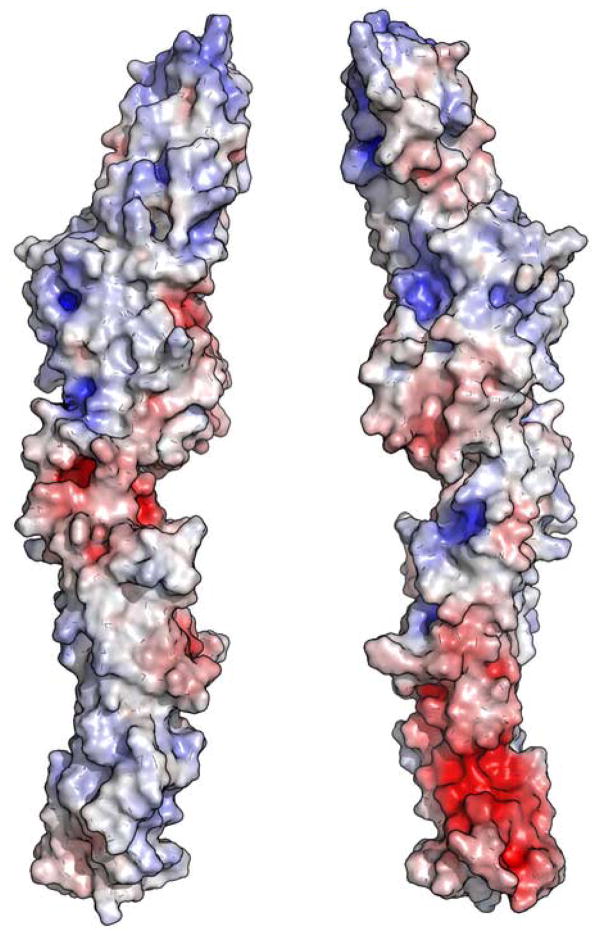
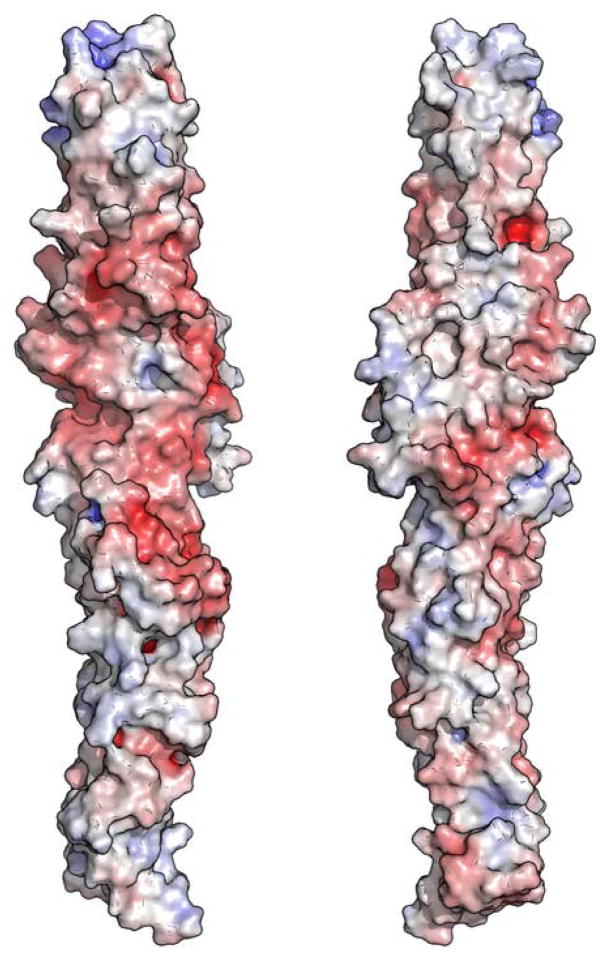
ScExo70 has been previously noted for the polarity of its surface electrostatic potential23,30. Its N-terminus is strongly electronegative, resulting from an electronegative patch composed mostly of residues from H2, H3 and H5 (Figure 6a). Its C-terminus is primarily electropositive, resulting mostly from residues on L10–11, H11, L13–14, H14, and H16-H19. This polarity is not observed in MmExo70 (Figure 6b). The N-terminus lacks the strong electronegative patch, and the C-terminus retains only a small electropositive patch at the extreme tip of the molecule. Despite this difference, the C-terminal basic patch remains a conserved feature on the surface of the molecule. In MmExo70 it is composed of Lys561, Arg563, Lys565, Arg567, Lys571, Lys575, Lys628, Lys632, and Lys635. All of these residues except Lys561, Arg563, and Lys575 are conserved, suggesting a common biological function. Interestingly, Lys571 and Lys628 have been implicated in Arpc1 interaction26. The middle of MmExo70 has an overall electronegative potential that contrasts with the mixed potential of ScExo70 in this region. Residues primarily from H4, L4–5, H7, H9-H12, H14-H15, L17–18 and H18 contribute to this patch. Only a few small regions remain unchanged between the two structures. H6, L16–17 and L18–19 compose two electropositive patches while H7 and H9, and H14-H15 compose two electronegative pockets. Therefore, it appears that Exo70 has highly variable surface electrostatic properties suggesting great variability in Exo70-mediated protein-protein interactions.
Figure 6. Surface electrostatic potential of Exo70.
The molecular surfaces of ScExo70 (A) and MmExo70 (B) are shown. The surfaces are colored on the basis of the solvent-accessible electrostatic potential of the molecules. Blue and red depict positive and negative electrostatic potential, respectively, with a range of ±10kBT/e. Views in the left panels are similar to Figure 1 and are related to the right panels by a 180° rotation about the long axis.
Similarity to other structures
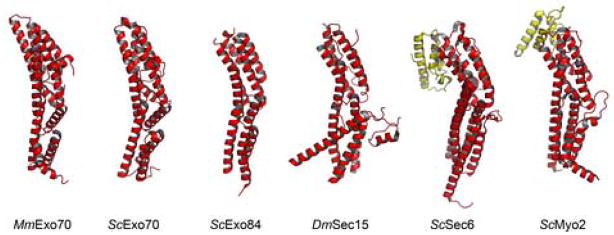
It has been noted that the N domain of ScExo7023, the C-terminal domain of S. cerevisiae Exo8423, the C-terminal domain of S. cerevisiae Sec6 (ScSec6)33 and the C-terminal domain of Drosophila melanogaster Sec15 (DmSec15)34 each share a similar fold33,35. Not surprisingly, the N domain of MmExo70 also contains this structural feature. A search of the RCSB Protein Data Bank36 for other proteins with a fold similar to the MmExo70 N domain using the DALI server37 also identifies the cargo-binding domain of S. cerevisiae Myo2 (ScMyo2), a Myosin V motor protein38 (Figure 7). This is the first instance of this fold appearing in a protein outside of the exocyst complex. The cargo-binding domain of ScMyo2 is located at the C-terminus of the molecule and contains an additional three-helix bundle at its C terminus, similar to ScSec6 and possibly DmSec1535. Notably, Rho3 regulates the functions of ScMyo2 and ScExo70, and the interaction of ScMyo2 with Rho3 results in exocytic vesicle delivery that functions alongside exocytosis12,28. We also note that of the five proteins now known to contain this fold, Exo70 is unique in having this domain at its N terminus. The function of this fold remains unclear despite its presence in these structures.
Figure 7. A common fold is found in Exo70, Myo2 and other structures.
Cartoon diagrams of molecules containing structures similar to the N domain of MmExo70. From left to right these are: MmExo70 N domain, ScExo70 N domain, ScExo84 C-terminal domain (PDB ID 2D2S), DmSec15 C-terminal domain (PDB ID 2A2F), ScSec6 C-terminal domains (PDB ID 2FJI), and ScMyo2 C-terminal domains (PDB ID 2F6H). Domains common in all these structures are colored red. ScSec6 and ScMyo2 terminate in a similar three-helix bundle (yellow) not seen in other structures.
TC10 binding
The structure of MmExo70 may reveal details important to understanding the interaction of mammalian Exo70 with small GTPase TC10 that are not available from that of ScExo70. In H. sapiens Exo70, residues 1–384 interact with TC10 while affinity is reduced with residues 1–99 and 100–384 individually19. The simplest explanation is that the site of interaction with TC10 spans both of these regions. The first 99 residues of MmExo70 extend nearly to the end of H1. Due to a lack of structural information for the first 84 residues of MmExo70, it is unclear how it might be involved in TC10 interaction, although secondary structure prediction suggests that these missing residues may also adopt a similar helix-turn-helix motif as observed in the rest of the molecule32. Residues 100–384 contain L4–5 and L6–7, both of which are longer in MmExo70 than in ScExo70. These significant structural differences could play important roles in mediating species-specific interactions with TC10, although it is equally possible that the strikingly different surface composition between these molecules could also play an important role.
Conclusion
The structure of MmExo70 reveals that, despite the conservation of the overall fold of Exo70, especially of the N domain that has structural similarity to other proteins involved in exocytosis, there are several major structural reorganizations that result in a different overall shape of the molecule. Packing within the M domain is altered and positions the C domain along the long axis of the molecule. A larger buried surface area and additional interactions add to the stability of the flexible hinge between N and M domains of ScExo70. A lack of sequence conservation for surface residues results in a drastic change in the surface electrostatic potential of the molecule. These differences in Exo70 structure are important to our understanding of many species-specific functions of the exocyst as more is learned about this fascinating molecular machinery.
MATERIALS AND METHODS
MmExo70 expression and purification
The MmExo70Δ84 gene (coding for residues 85–653) was amplified from M. musculus cDNA (provided by A. Saltiel) by PCR and subcloned into the pSJ7 vector, a derivative of pET43a (Novagen). The native protein was expressed in Escherichia coli BL21(DE3) strain containing the expression plasmid at 16°C in LB media, and the L-Selenomethionine variant protein was expressed in Escherichia coli B834(DE3) strain containing the expression plasmid at 16°C in L-Selenomethionine-containing minimal media. Cells were induced with 0.4mM IPTG (Calbiochem) and protein purification was carried out at 4°C. Cell pellets were lysed by sonication in buffer A (50mM Tris pH8.0, 300mM NaCl, 5mM β-mercaptoethanol, 10μg/ml PMSF), and soluble material was passed over a Ni2+-NTA column and eluted with a linear gradient of buffer B (buffer A with 250mM imidazole). The N-terminal NusA-His-tag was removed by proteolysis with TEV protease during dialysis against dialysis buffer (50mM Tris pH8.0, 100mM NaCl, 5mM β-mercaptoethanol, 10μg/ml PMSF). TEV protease and the tag were removed by passage over a second Ni2+-NTA column. MmExo70 was further purified on a SourceQ column, loaded in buffer C (50mM Tris pH8.0, 1mM dithiothreitol) and eluted with a linear gradient of buffer D (buffer C with 1M NaCl). Purified protein was stored in crystallization buffer (50mM Tris pH8.0, 100mM NaCl, 1mM Tris[2-carboxyethyl]phosphine) at a concentration of 14.5mg/mL (native) or 13.5mg/mL (L-selenomethionine variant).
MmExo70 crystallization and data collection
Native or L-Selenomethionine variant crystals appeared in 1–2 days and reached full size in 7–10 days at 20°C using the sitting drop method. Equal volumes of protein and precipitant (100mM HEPES pH7.5, 10% ethylene glycol, 5% PEG8000, 10mM MgCl2) were mixed in a 4μL drop and equilibrated over 500μL of well solution (100mM HEPES pH7.5, 10% ethylene glycol, 8% PEG8000). Streak seeding was performed to improve the quality of crystals. Formation of a sticky film with time necessitated prompt harvesting. Crystals were washed in cryo-solution (100mM HEPES pH7.5, 30% ethylene glycol, 10% PEG8000, 10mM MgCl2) before being flash cooled in liquid nitrogen. Native and MAD data were collected at beamline 23-D at the Advanced Photon Source and were processed using HKL200039.
MmExo70 refinement and model building
Experimental phases were obtained using the multi-wavelength anomalous dispersion (MAD) method40. Initial heavy atom sites were found using CNS41 and confirmed using Shake-N-Bake42. Refinement of the heavy atom sites was carried out using autoSHARP43, and ten of the expected eleven selenium sites in the monomer were found (the missing selenium site is located in a disordered portion of the molecule). MAD phases were calculated, solvent flattening was performed, and the initial model was built using autoSHARP. The remainder of the model was built based on the locations of known selenium atoms using O44, and all refinement procedures were carried out using CNS. Initial refinement consisted of several iterations of simulated annealing by the maximum likelihood target function using amplitudes and phase probability distribution (MLHL), grouped B factor refinement, and model rebuilding using O. This was done using data from the peak wavelength of the MAD data set. Further refinement was done against the native data set to its limiting resolution of 2.25Å. This consisted of iterations of simulated annealing by the maximum likelihood target function using amplitudes, individual B factor refinement, and model building using O. 3Fo-2Fc and Fo-Fc maps were calculated to aid model building and water placement. The final model consists of residues 85–179, 188–241, 275–446, 455–652, and 170 water molecules.
Figure preparation
Figures 1, 3, 4, 5, and 7 were produced with MacPyMOL (http://www.pymol.org). The sequence alignment shown in Figure 2 is based on structural alignment of MmExo70 with ScExo70. Both structures were broken into a series of overlapping three-helix fragments, which were aligned by the DALI server45. The sequences of Exo70 from Candida albicans, S. pombe, Caenorhabditis elegans, D. melanogaster, and H. sapiens were aligned with the MmExo70/ScExo70 alignment based on Clustal W46. Residues invariable in six of these seven species were considered completely conserved. Residues that always appear as a particular residue type in six of these seven species were considered similarly conserved. Figure 6 was produced using the adaptive Poisson-Boltzmann solver47 in the APBS Tools plug-in for PyMOL (www.umich.edu/~mlerner/Pymol). All missing side chains, but not completely missing residues, were modeled in a rotamer allowed by the surrounding structure for this calculation. Structural alignment in Figures 5, 6, and 7 were performed using the DALI server or PyMOL.
Protein Data Bank accession number
Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank with accession code 2PFT for MmExo70 and 2PFV for ScExo70.
Supplementary Material
Acknowledgments
We thank the staffs of the Center for Structural Biology at the University of Michigan for maintaining the X-ray facility; the staffs at the APS GM/CA-CAT beamline in the Argonne National Laboratory for access and help with data collection; and Alan R. Saltiel, Janet L. Smith, and John Tesmer for critical reading of the manuscript. B.M. was a trainee in the Molecular Mechanisms in Microbial Pathogenesis training program at the University of Michigan Medical School. This work was supported in part by NIH grants to Z.X. (R01-DK65980).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnston GC, Prendergast JA, Singer RA. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. Embo J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 4.Folsch H, Pypaert M, Maday S, Pelletier L, Mellman I. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J Cell Biol. 2003;163:351–362. doi: 10.1083/jcb.200309020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prigent M, Dubois T, Raposo G, Derrien V, Tenza D, Rosse C, Camonis J, Chavrier P. ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J Cell Biol. 2003;163:1111–1121. doi: 10.1083/jcb.200305029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ang AL, Taguchi T, Francis S, Folsch H, Murrells LJ, Pypaert M, Warren G, Mellman I. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J Cell Biol. 2004;167:531–543. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo W, Grant A, Novick P. Exo84p is an exocyst protein essential for secretion. J Biol Chem. 1999;274:23558–23564. doi: 10.1074/jbc.274.33.23558. [DOI] [PubMed] [Google Scholar]
- 9.Hsu SC, Ting AE, Hazuka CD, Davanger S, Kenny JW, Kee Y, Scheller RH. The mammalian brain rsec6/8 complex. Neuron. 1996;17:1209–1219. doi: 10.1016/s0896-6273(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 10.Kee Y, Yoo JS, Hazuka CD, Peterson KE, Hsu SC, Scheller RH. Subunit structure of the mammalian exocyst complex. Proc Natl Acad Sci U S A. 1997;94:14438–14443. doi: 10.1073/pnas.94.26.14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo W, Tamanoi F, Novick P. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat Cell Biol. 2001;3:353–360. doi: 10.1038/35070029. [DOI] [PubMed] [Google Scholar]
- 12.Robinson NG, Guo L, Imai J, Toh EA, Matsui Y, Tamanoi F. Rho3 of Saccharomyces cerevisiae, which regulates the actin cytoskeleton and exocytosis, is a GTPase which interacts with Myo2 and Exo70. Mol Cell Biol. 1999;19:3580–3587. doi: 10.1128/mcb.19.5.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Bi E, Novick P, Du L, Kozminski KG, Lipschutz JH, Guo W. Cdc42 interacts with the exocyst and regulates polarized secretion. J Biol Chem. 2001;276:46745–46750. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- 14.Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. Embo J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin R, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. Embo J. 2005;24:2064–2074. doi: 10.1038/sj.emboj.7600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. The exocyst is a Ral effector complex. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- 17.Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, Camonis J, White MA. Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- 18.Sugihara K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y. The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat Cell Biol. 2002;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- 19.Inoue M, Chang L, Hwang J, Chiang SH, Saltiel AR. The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature. 2003;422:629–633. doi: 10.1038/nature01533. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XM, Ellis S, Sriratana A, Mitchell CA, Rowe T. Sec15 is an effector for the Rab11 GTPase in mammalian cells. J Biol Chem. 2004;279:43027–43034. doi: 10.1074/jbc.M402264200. [DOI] [PubMed] [Google Scholar]
- 21.Grote E, Carr CM, Novick PJ. Ordering the final events in yeast exocytosis. J Cell Biol. 2000;151:439–452. doi: 10.1083/jcb.151.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walch-Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat Struct Mol Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- 24.Matern HT, Yeaman C, Nelson WJ, Scheller RH. The Sec6/8 complex in mammalian cells: characterization of mammalian Sec3, subunit interactions, and expression of subunits in polarized cells. Proc Natl Acad Sci U S A. 2001;98:9648–9653. doi: 10.1073/pnas.171317898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vega IE, Hsu SC. The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J Neurosci. 2001;21:3839–3848. doi: 10.1523/JNEUROSCI.21-11-03839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuo X, Zhang J, Zhang Y, Hsu SC, Zhou D, Guo W. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat Cell Biol. 2006;8:1383–1388. doi: 10.1038/ncb1505. [DOI] [PubMed] [Google Scholar]
- 27.Inoue M, Chiang SH, Chang L, Chen XW, Saltiel AR. Compartmentalization of the exocyst complex in lipid rafts controls Glut4 vesicle tethering. Mol Biol Cell. 2006;17:2303–2311. doi: 10.1091/mbc.E06-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adamo JE, Rossi G, Brennwald P. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol Biol Cell. 1999;10:4121–4133. doi: 10.1091/mbc.10.12.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyd C, Hughes T, Pypaert M, Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamburger ZA, Hamburger AE, West AP, Jr, Weis WI. Crystal structure of the S. cerevisiae exocyst component Exo70p. J Mol Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- 31.Ramakrishnan C, Ramachandran GN. Stereochemical criteria for polypeptide and protein chain conformations. II. Allowed conformations for a pair of peptide units. Biophysical Journal. 1965;5:909–933. doi: 10.1016/S0006-3495(65)86759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 33.Sivaram MV, Furgason ML, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat Struct Mol Biol. 2006;13:555–556. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- 34.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 35.Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 38.Pashkova N, Jin Y, Ramaswamy S, Weisman LS. Structural basis for myosin V discrimination between distinct cargoes. Embo J. 2006;25:693–700. doi: 10.1038/sj.emboj.7600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Hendrickson WA, Ogata CM. Phase Determination from Multiwavelength Anomalous Diffraction Measurements. Methods in Enzymology. 1997;276:494–523. doi: 10.1016/S0076-6879(97)76074-9. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Weeks CM, Miller R. Optimizing Shake-and-Bake for proteins. Acta Crystallogr D Biol Crystallogr. 1999;55:492–500. doi: 10.1107/s0907444998012633. [DOI] [PubMed] [Google Scholar]
- 43.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated Structure Solution With autoSHARP. Methods Mol Biol. 2006;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 44.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 (Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 45.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.