Fig. 5.
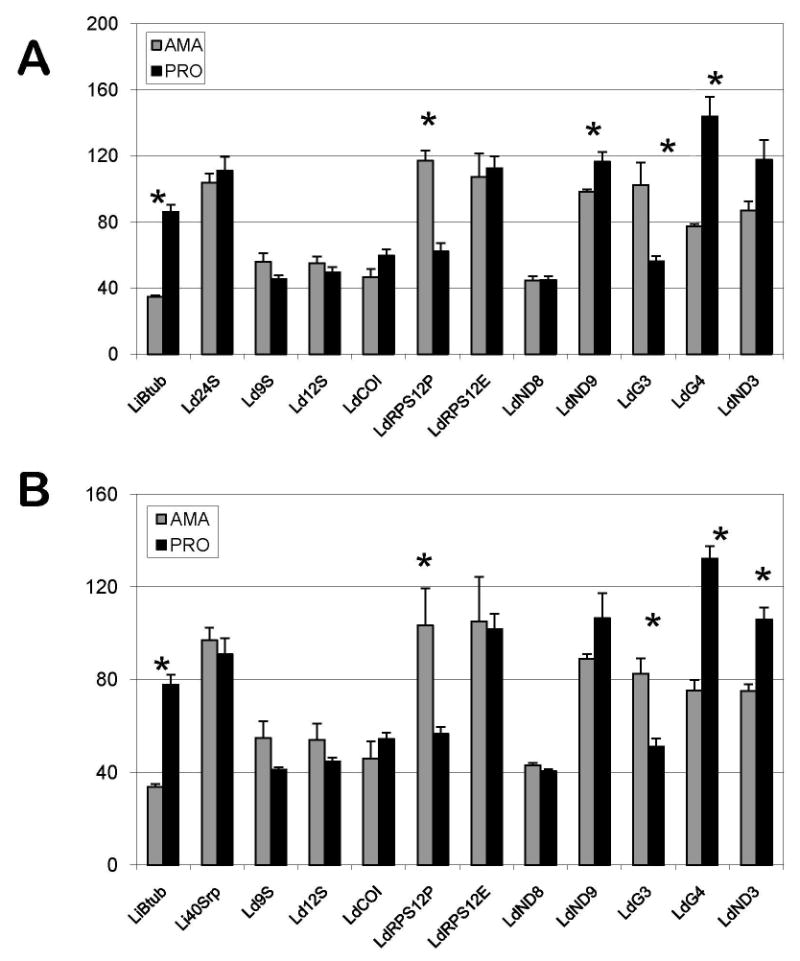
Quantitative reverse transcriptase-PCR analysis of nuclear and mitochondrial transcripts in amastigotes and promastigotes of Leishmania donovani. The columns represent averages of three replicates normalized to mRNA of Leishmania infantum cytoplasmic 40S subunit ribosomal protein S12 (A), designated Li40Srp, and L. donovani 24Sa ribosomal RNA (B), designated Ld24S. Other designations for L. donovani transcripts are: Ld9S and Ld12S - mitochondrial ribosomal 9S and 12S RNA, respectively; LdRPS12P and LdRPS12E - pre-edited and edited mRNA, respectively, of mitochondrial ribosomal protein S12; LdCOI - cytochrome c oxidase subunit I mRNA; LdND3, LdND8 and LdND9 -subunits 3, 8 and 9 of NADH dehydrogenase mRNA; LdG3 and LdG4 - pan-edited cryptogenes G3 and G4 mRNA. LiBtub - L. infantum (β-tubulin mRNA. The vertical axis shows the relative abundance of the respective mRNAs in amastigotes (grey) and promastigotes (black) with a standard error as shown. The asterisks positioned above some columns (β-tubulin, pre-edited mitochondrial RPS12, ND9, G3, G4 and ND3 transcripts) indicate that the observed differences in relative amount statistically significant (with P < 0.05, derived from Student's test).
