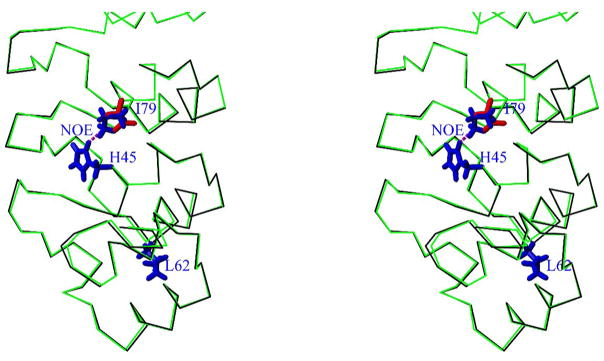
Figure 10. Stereoview showing the overlay of Cα trace of gankyrin WT (black) and modeled I79D mutant (green).
Only the AR1-AR3 repeats are shown. The side chains of the following residues are highlighted: H45, L62 and I79 of gankyrin WT (in blue), and D79 of I79D mutant (in red). In previous NMR studies an NOE between Hδ2/H45 and Hδ1/I79 was observed (dashed line in magenta) 21. Apparently the modeling I79D structure indicates that such side-chain interactions would be impossible without major structural adjustment. Thus the local structures around H45 including the binding residues may be destabilized by this mutation.