Figure 1. Two-way hierarchical clustering of expression data.
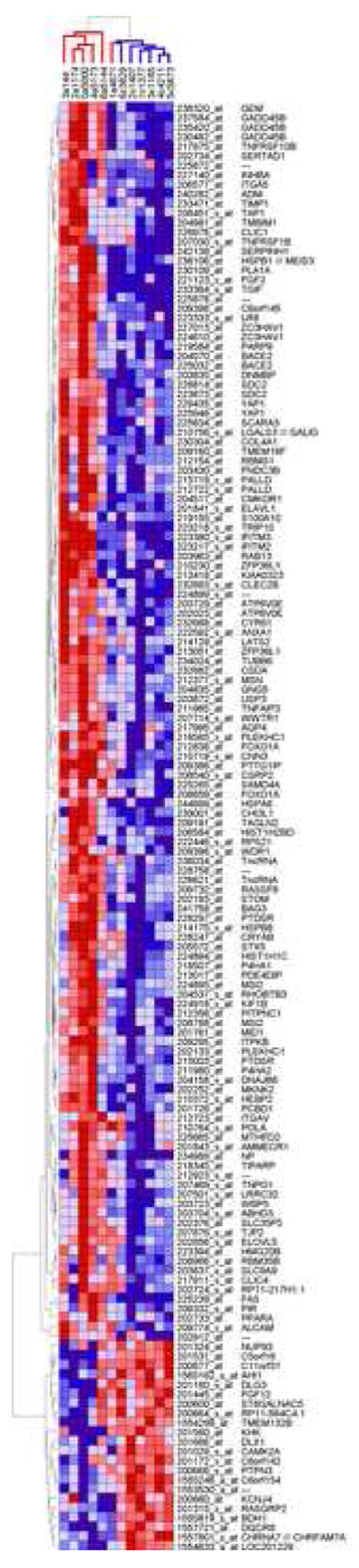
Hierarchical clustering was performed on log2-transformed expression level of 152 differentially expressed genes which demonstrated |ALR|>1 and p<0.05 in three distinct statistical analyses (groupwise, pairwise and rank-based). Samples were clustered vertically, gene probes were clustered horizontally. Genes are denoted by Affymetrix probes and NCBI gene symbols. Each colored square represents a normalized gene expression level, color coded for increase (red) or decrease (blue). Color intensity is proportional to magnitude of change. The clustering resulted in a near-perfect separation of samples into two discrete groups corresponding to diagnosis (vertical dendrogram, red for AUT and blue for CONT).
