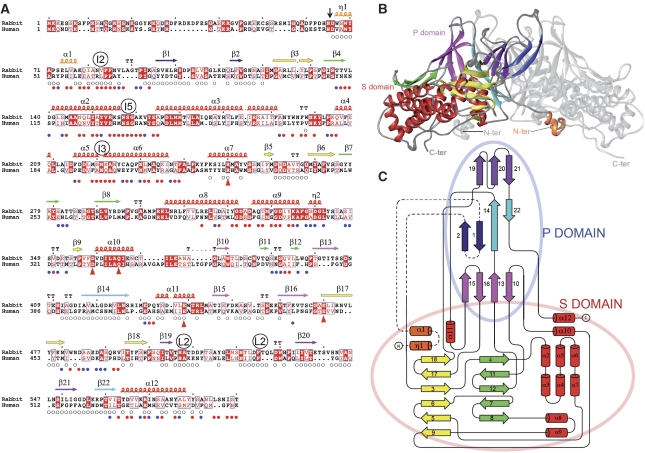
Figure 1.
Structure of the PBV CP. (A) Amino-acid sequence alignment of rabbit and human PBV CP (accession Q9Q1V2 and Q50LE5, respectively) highlighting conserved amino acids. A vertical arrow points to the N-terminal residue of the 55 kD CP found in the particle. Secondary structure elements (SSE) are labelled as α, η and β, for α-helices, 3/10-helices and β-strands, respectively, numbered sequentially from the N terminus and coloured to match the diagrams of the other panels. A labeled circle above the sequence marks residues approaching the icosahedral (I2, I3 and I5) and local (L2) symmetry axes. Small circles below the sequence flag residues involved in L2-related intra-dimer contacts (open circles) and between dimers in the capsid (full coloured circles: blue and red denote single and double (meaning two non-equivalent) contacts in the capsid, respectively. Red triangles below the sequence flag residues involved in the hydrogen-bonding network surrounding the transproteolytic cleavage site. (B) Ribbon diagram of the CP dimer. Shell and Projecting domains are labelled S and P, and the N- and C-termini are indicated. One subunit is coloured by SSE, with all helices in red except for the swapped region (η1-α1, orange) and β-sheets in yellow and green in the S domain, and purple, blue and pink in the P domain. In the blue sheet, dark blue indicates the swapped, N-terminal β1–β2 hairpin. For clarity, the second subunit is shown in grey. (C) Topology diagram. β-strands and α or η helices are indicated by arrows and cylinders, respectively, coloured and numbered as in A, and connected by a black line. A broken line connects elements that are swapped between the two subunits in the dimer. Pale red and blue ovals encompass secondary structure elements of the S and P domains, respectively.