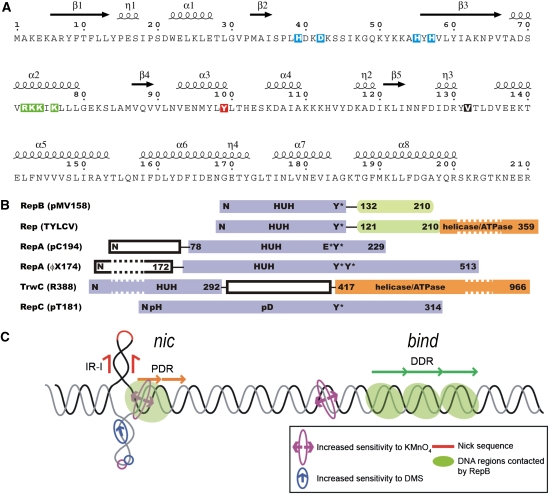
Figure 1.
Secondary structure of RepB, domain organization of RepB and related proteins, and dso of pMV158. (A) The primary sequence of RepB, including the annotation of the secondary structure elements as found in the C2- and C3-structures, η denotes 310-helices. The residues involved in metal coordination, which include the histidines of the HUH sequence motif (residues H55, Y56 and H57), are boxed in blue. The catalytic tyrosine is boxed in red, the mutated residues are boxed in green and hinge V132 is shown as white on a black background. (B) Alignment of the tyrosine residues responsible for nicking activity in various RCR initiator proteins (adapted from Campos-Olivas et al, 2002). The catalytic tyrosine of TrwC, which is at the N-terminus due to a circular permutation in the sequence, cannot be aligned with the others. The reactive tyrosines are marked with Y*, and metal-binding sequence motifs are marked with ‘HUH'. Several poly His (pH) and poly Asp (pD) sequences were identified in the sequence of RepC from plasmid pT181, which could be involved in metal binding. Some of the N-terminal and C-terminal domains have been shortened (indicated by dashed boxes). (C) Schematic representation of the pMV158 dso, showing the approximate relative locations of the nick site, the proximal direct repeats (PDR) of the nic locus and the distal direct repeats (DDR) constituting the bind locus. The regions that are contacted by RepB and the regions with increased sensitivity to KMnO4 or dimethyl sulphate (DMS) on binding of RepB are also indicated.