Abstract
Activated antigen-specific T cells produce a variety of effector molecules for clearing infection, but also contribute significantly to inflammation and tissue injury. Here we report an anti-inflammatory property of anti-viral CD8+ and CD4+ effector T cells (Te) in the infected periphery during acute virus infection. We find that, during acute influenza infection, IL-10 is produced in the infected lungs at high levels -- exclusively by infiltrating virus-specific Te, with CD8+ Te contributing a larger fraction of the IL-10 produced. These Te in the periphery simultaneously produce IL-10 and proinflammatory cytokines, and express lineage markers characteristic of conventional Th/c1 cells. Importantly, blocking the action of the Te-derived IL-10 results in enhanced pulmonary inflammation and lethal injury. Our results demonstrate that anti-viral Te exert regulatory functions -- that is, fine-tune the extent of lung inflammation and injury associated with influenza infection by the production of an anti-inflammatory cytokine. The potential implications of these findings for infection with highly pathogenic influenza viruses are discussed.
INTRODUCTION
The innate and adaptive immune systems play pivotal roles in orchestrating the host response to infection and tissue injury. The host responses to such insults include the production of a variety of pro-inflammatory cytokines and chemokines. The induction of the pro-inflammatory response triggers the development of a counter-regulatory anti-inflammatory cytokine response to control inflammation and prevent excessive injury. In certain instances (e.g. infection with the highly pathogenic avian H5N1 or 1918 pandemic influenza virus strains), this counter regulation fails since infection results in a massive inflammatory cell infiltration into the infected lungs and excessive pro-inflammatory cytokine production 1–3.
IL-10 has been recognized as an important anti-inflammatory cytokine serving as a negative regulator of the response of both innate and adaptive immune cells particularly during persistent bacterial and parasitic infections where it suppresses pathogen clearance and/or infection associated immunopathology 4,5. More recently, IL-10 has been recognized to mediate virus persistence during chronic viral infections 6–8. Multiple cell types have been reported to produce IL-10 either constitutively or in response to inflammatory stimuli, most notably “regulatory” T-cells and dendritic cells and recently CD4+ effector T-cells during protozoa infections 4,9–12. IL-10 has not been reported to play any significant role in acute virus infections.
Here, we analyzed the production, cellular source and function of IL-10 during acute respiratory influenza virus infection. Surprisingly, adaptive immune CD8+ and CD4+ effector T-cells (Te) were the primary source of IL-10 produced in the influenza infected respiratory tract with CD8+ Te contributing larger fraction of the total IL-10 produced. IL-10 and pro-inflammatory cytokines were simultaneously produced by these Te early in the adaptive response. Blockade of the action of IL-10 in vivo in sub-lethally infected animals resulted in enhanced pulmonary inflammation (elevated inflammatory cell infiltration and cytokine and chemokine production), lethal injury and accelerated death with no effect on the tempo of virus clearance. Lethal injury was partially reversed by corticosteroid administration. Our findings suggest that Te-derived IL-10 may play a critical role in regulating the magnitude of inflammation during acute virus infection.
RESULTS
Influenza-specific T cells preferentially produce IL-10 and IFN-γ in the infected lungs
We analyzed cytokine secretion by CD4+ and CD8+ T cells infiltrating the lungs of influenza A/PR/8-infected mice. Unexpectedly, we found that a significant fraction of both CD4+ and CD8+ T cells from the infected lungs secreted IL-10 and/or IFN-γ (and minimal IL-4 or IL-17) in response to PMA/ionomycin stimulation in the intracellular cytokine synthesis (ICS) assay (Supplementary Fig. 1 online). Furthermore, the mitogen-induced IL-10 synthesis was restricted to Thy-1+ lymphocytes in the lungs (Supplementary Fig. 2 online).
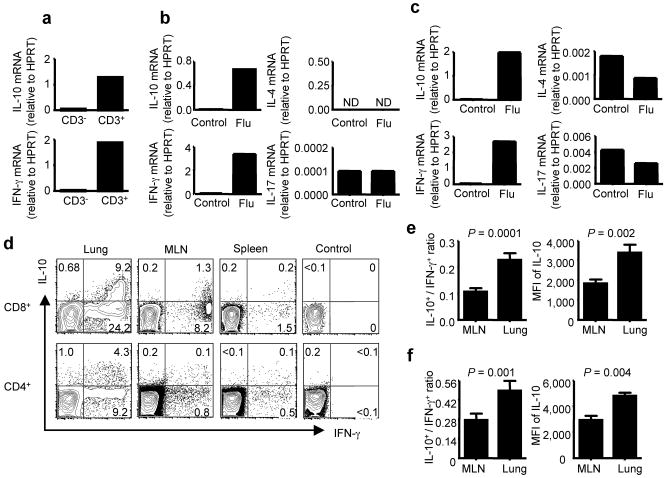
We next compared the levels of IL-10 and IFN-γ transcripts in purified CD3+ cells and the residual CD3− cells isolated from influenza-infected lungs. We observed that, like IFN-γ message, the expression of IL-10 transcripts was primarily restricted to this CD3+ cell fraction (Fig. 1a). We also detected the expression of IL-10 mRNA in both purified CD8+ (Fig. 1b) and CD4+ (Fig. 1c) T cells isolated from the infected lungs while minimal levels of IL-4 and IL-17 mRNA were detected in either T cell subsets. To determine whether lung T cells are capable of producing IL-10 in response to viral antigen, we used ICS assay to examine IL-10 and IFN-γ production by CD8+ and CD4+ T cells isolated from lungs, the draining mediastinal lymph nodes (MLN) and spleens of day 8 infected mice following stimulation with influenza-infected BMDC. We found that a significant fraction of CD8+ and CD4+ T cells of the infected lungs and a smaller fraction of CD8+ and CD4+ T cells in MLN and spleens were capable of producing IL-10 in response to viral antigen (Fig. 1d) with a higher percentage of antigen-specific CD8+ and CD4+ T cells (as indicated by the IFN-γ production) in the lungs capable of producing IL-10 than those detected in the MLN (Fig. 1e, f). Furthermore, the IL-10-producing T cells in the lungs expressed higher levels of IL-10 than the corresponding cells in the MLN (Fig. 1e, f). We also detected the induction of IL-10 producing CD8+ T cells with recall response to heterologous influenza virus challenge and during primary RSV infection (Supplementary Fig. 3 online and data not shown).
Figure 1. Influenza-specific T cells preferentially produce IL-10 and IFN-γ in the influenza-infected BALB/c lungs.
(a) Quantitative RT-RCR analysis of IL-10 and IFN-γ transcript levels of purified CD3+and CD3− cells in the lungs of mice 8 days p.i. (b, c) Quantitative RT-RCR analysis of IL-4, IL-17, IL-10 and IFN-γ transcript levels of purified CD8+(b) or CD4+(c) T cells in the lungs of mice 8 days p.i. (d) At day 8 p.i., lung, mediastinal lymph node (MLN) and spleen cells were restimulated in vitro with influenza infected BMDC and intracellular cytokine staining (ICS) was performed to measure the production of IL-10 and IFN-γ in gated CD8+ and CD4+ T cells. Numbers indicate the percentages of cytokine-positive cells in the gates within the total population. (e, f) Cells from MLN and lungs of mice 8 days p.i. were restimulated with flu-infected BMDC and ICS was performed to measure the production of IL-10 and IFN-γ in gated CD8+ (e) and CD4+ (f) T cells. The ratio of IL-10+ vs. IFN-γ+ CD8+ (e) or CD4+ (f) T cells and the mean fluorescence intensity (MFI) of IL-10 staining in IL-10+ CD8+ (e) or IL-10+ CD4+ (f) T cells in MLN and lungs are depicted. P values were determined by unpaired two-tailed student t test.
IL-10-producing CD8+ T cells and most IL-10-producing CD4+ T-cells are virus-specific effector T cells (Te)
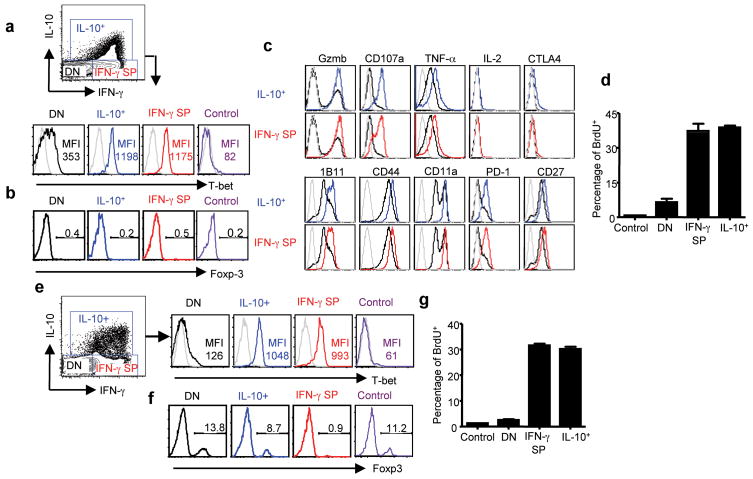
IL-10 production has typically been detected in CD4+ effector and regulatory T-cells 9. Our observation (i.e. that a significant fraction of antigen-specific CD8+ T cells at the site of infection in the lung were capable of responding to antigen with IL-10 production) was unexpected and to our knowledge, has not been previously demonstrated. To determine the phenotype of these IL-10 producing (IL-10+) CD8+ T cells, we first examined the expression of T-bet and Foxp-3 by the IL-10+ CD8+ T cells, the “conventional” CD8+ IFN-γ single positive Tc1 Te (IFN-γ SP) and CD8+ IL-10− IFN-γ− (DN) T cells. Like Tc1 Te, these IL-10+ T cells expressed high levels of T-bet but not the regulatory cell marker Foxp-3 (Fig. 2a, b). Further confirmation that these IL-10-producing T cells were Tc1 Te came from the demonstration of the expression by these IL-10+ T-cells of effector molecules (e.g. Granzyme B, CD107a, TNF-α, etc.) characteristic of conventional Tc1 Te (Fig. 2c). Moreover IL-10-producing CD8+ T cells were also capable of proliferating on site in the infected lungs as measured by a 2-hour BrdU incorporation assay (Fig. 2d), suggesting that, in contrast to IL-10 producing CD4+ Tr1 cells 13, they were not anergic in vivo. We also established that the majority of the IL-10 producing CD4+ T cells had a phenotypic profile characteristic of effector Th1 Te (Fig. 2e, f), although a very small fraction of IL-10 producing CD4+ T cells co-expressed T-bet and Foxp-3 and thus exhibited the phenotype of the regulatory T cells described before 14 (Fig. 2e, f). Like the IL-10 producing CD8+ Te, IL-10 producing CD4+ T cells were capable of proliferating in situ in the infected lungs (Fig. 2g).
Figure 2. IL-10 producing CD8+ and CD4+ T cells are Type 1 effectors.
BALB/c mice were infected with influenza. CD8+ (a–c) or CD4+ (e, f) cells from lungs of uninfected mice or mice 7 days p.i. with influenza were restimulated with influenza infected BMDC and the expression of T-bet (a, e), Foxp-3 (b, f) and a panel of effector or activation molecules (c) in uninfected lung CD8+ or CD4+ T cells (control) or IL-10 positive CD8+ or CD4+ T cells (IL-10+), IFN-γ single positive CD8+ or CD4+ T cells (IFN-γ SP), IL-10 and IFN-γ double negative CD8+ or CD4+ T cells (DN) from infected lungs were measured by ICS. Gray line: isotype control Ab staining. Numbers are MFI (a, e) or percentages of Foxp-3-positive cells in the gates within the total population (b, f). (d, g) Uninfected mice (control) and mice 6 days p.i. were injected with BrdU. 2 h later, mice were sacrificed and lung cells were restimulated with influenza-infected BMDC. The percentages of BrdU+ cells in CD8+ (d) or CD4+ (g) cells of uninfected lung (control) and the IL-10+, IFN-γ single positive (IFN-γ SP) and IL-10− IFN-γ− (DN) T cells of 6 days p.i. lung are depicted.
IL-10 is expressed by both CD8+ and CD4+ Te, but only by memory CD4+ T cells
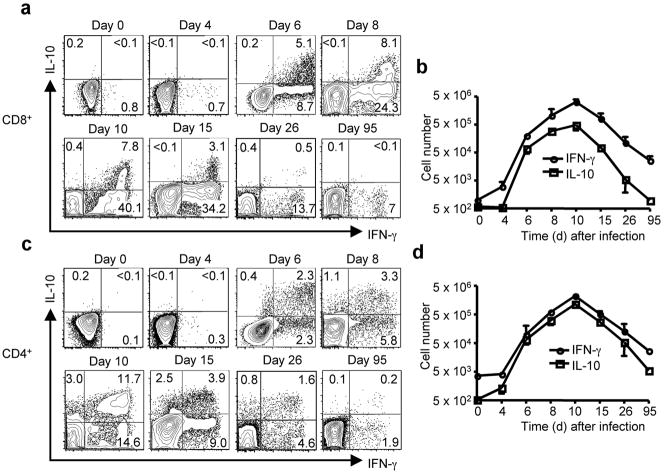
We next examined the kinetcs of expression of IL-10 by virus-specific CD8+ and CD4+ T cells in infected lungs. Consistent with the timing of CD8+ Te migration during influenza infection, IL-10+ lung CD8+ Te were first readily demonstrable at day 6 p.i. (Fig. 3a). The total number of IL-10+ and IFN-γ+ CD8+ Te in the lungs continued to increase until day 10 p.i.; and CD8+ Te capable of secreting IL-10 were still detectable during the early contraction phase of the response (i.e. day 15) (Fig. 3b). However, with additional contraction of the response and the transition into the memory phase (i.e., day 26 and beyond), the fraction of IL-10-producing T cells in the remaining virus-specific (IFN-γ+) CD8+ T-cells was markedly decreased and eventually became undetectable (Fig. 3a, b).
Figure 3. Kinetics of IL-10 producing CD8+ and CD4+ Te in vivo.
BALB/c mice were infected with influenza. At the indicated days p.i, lung cells were restimulated with influenza-infected BMDC. The production of IL-10 and IFN-γ by CD8+ and CD4+ T cells was measured by ICS. (a, c) FACS plots of IL-10 and IFN-γ production by CD8+ (a) or CD4+ (c) T cells in the lung Numbers indicate the percentages of cytokine-positive cells in the gates within the CD8+ (a) or CD4+ (c) T cell population. (b, d) The kinetics of accumulation (absolute numbers) of IL-10+ or IFN-γ+ CD8+ T cells (b) or IL-10+ or IFN-γ+ CD4+ T cells (d) in the lungs following influenza infection.
The kinetics of accumulation of IL-10-expressing CD4+ Te into infected lungs directly paralleled that of IL-10 producing CD8+ Te (Fig. 3c, d). IL-10 production was largely restricted to IFN-γ+ CD4+ Te, but, in contrast to CD8+ Te, a fraction of CD4+ Te were IL- 10 “single positive” (Fig. 3c). In a further contrast with CD8+ Te, virus-specific IL-10+ memory CD4+ T cells were readily detected at day 26, and remained detectable as late as day 95 (Fig. 3c, d).
Te produce IL-10 in vivo during acute influenza infection
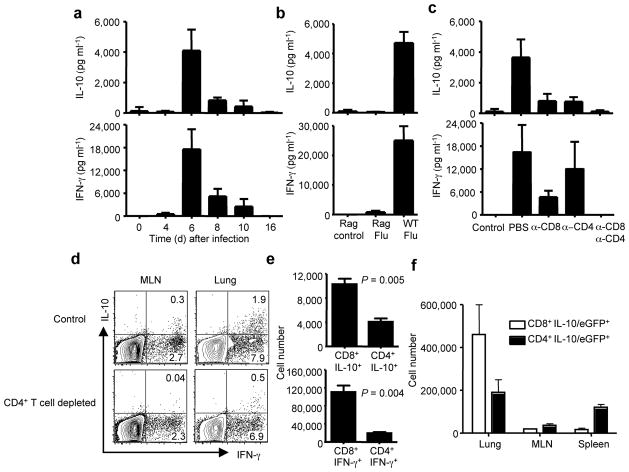
To determine if IL-10 was produced in vivo, we measured the kinetics of IL-10 and IFN-γ release into bronchial alveolar lavage fluid (BALF) sampled from infected lungs by ELISA. We found (Fig. 4a) that minimal levels of either cytokine were detected in the BALF early during infection (i.e. day 4 p.i.) in spite of high lung virus titers at this time 15. By day 6 p.i., both IL-10 and IFN-γ levels in the BALF increased dramatically and in a coordinated manner, with BALF levels of these two cytokines decreasing progressively through day 8 to 10 p.i., and returning to background levels by day 16. It is particularly noteworthy that this dramatic rise in the concentrations of these two cytokines at day 6 p.i. coincided with the recruitment of CD8+ and CD4+ Te into the infected lungs 16. Furthermore, the subsequent decline in the production of these two cytokines coincided with the fall in lung virus titers (and therefore viral antigen load) 17 even though the absolute number of CD8+ and CD4+ Te continued to increase in the lungs up to day 10 p.i. (Fig. 3b, d).
Figure 4. Te are the major source of IL-10 in vivo during influenza infection.
BALB/c mice were infected with influenza (a–e). (a) At the indicated days p.i., IL-10 and IFN-γ levels in the BALF were measured by ELISA. (b) WT and Rag1−/− mice were infected with influenza. At day 6 p.i., IL-10 and IFN-γ levels in the BALF were measured by ELISA. Rag control (Rag1−/− mice uninfected), Rag Flu (Rag1−/− mice infected with influenza), WT Flu (WT mice infected with influenza). (c) At day 6 p.i., IL- 10 and IFN-γ levels in the BALF were measured by ELISA. Control (uninfected mice), PBS (mice injected with PBS), α-CD8 (mice injected with CD8-specific mAb), α-CD4 (mice injected with CD4-specific mAb), α-CD8/α-CD4 (mice injected with CD8-specific plus CD4-specific mAbs). (d) At day 6 p.i., lung and MLN cells from control or CD4+ T cell depleted mice were restimulated in vitro with influenza-infected BMDC and ICS was performed to measure the production of IL-10 and IFN-γ in gated CD8+ T cells. Numbers indicate the percentages of cytokine-positive cells in the gates within the total population. (e) At day 6 p.i., mice were injected with monensin and the in vivo ICS assay was performed to measure the production of IL-10 and IFN-γ by CD8+ and CD4+ T cells in the lung in vivo. The numbers of IL-10+ CD8+, IL-10+ CD4+, IFN-γ+ CD8+ and IFN-γ+ CD4+ T cells are depicted. P values were determined by unpaired two-tailed student t test. (f) IL-10/eGFP reporter mice were infected with influenza. At day 6 p.i., the numbers of IL-10/eGFP+ CD8+ and IL-10/eGFP+ CD4+ T cells in the lungs, MLN and spleens are depicted.
In support of the hypothesis that infiltrating Te were the main IL-10 producer in vivo, we found that minimal levels of IL-10 and IFN-γ were produced in the BALF sampled from influenza infected Rag1−/− mice (Fig. 4b). Further confirmation of this hypothesis came from the analysis of IL-10 and IFN-γ levels in the mice after selective depletion of CD8+ and/or CD4+ T cells. The depletion of the CD8+ T cells substantially reduced IL-10 as well as IFN-γ release into the BALF, suggesting that CD8+ Te were an important source of the two cytokines in vivo (Fig. 4c). The depletion of CD4 T cells only slightly influenced the production of IFN-γ, but similarly, significantly inhibited the production of IL-10 in BALF, suggesting that IL-10 production in vivo was also dependent on CD4+ T cells (Fig. 4c). IL-10 and IFN-γ release into the BALF was reduced to control levels by the simultaneous depletion of CD8+ and CD4+ T cells (Fig. 4c). We were intrigued by the findings that lung IL-10 production was dependent on both CD8+ and CD4+ T cells during influenza infection in vivo. To investigate this phenomenon, we measured the induction of IL-10 producing CD8+ Te in vivo after the depletion of CD4+ T cells. We found that the CD4+ T cell depletion impaired the development of IL-10 producing CD8+ Te, but not IFN-γ or TNF-α producing Te (Fig. 4d and data not shown). Importantly, the induction of IL-10 producing CD4+ Te was independent of CD8+ T cells (Supplementary Fig. 4 online). To directly delineate the contribution of CD4+ and CD8+ Te in IL-10 production in vivo, we performed the in vivo ICS assay and found that the numbers of IL-10 producing CD8+ Te were 2–3 folds higher than IL-10 producing CD4+ Te in the lung (Fig. 4e and Supplementary Fig. 5 online), suggesting that CD8+ Te were a larger contributor to the Te-derived IL-10 in the infected lungs than CD4+ Te. We also monitored the induction of IL-10 producing cells in the lung following influenza infection of IL-10/eGFP reporter (Vert-X) mice. In these mice, IL-10/eGFP+ cells in infected lungs were tightly restricted to the Thy1+ T-lymphocyte population (Supplementary Fig. 6 online). Furthermore, although CD4+ IL-10/eGFP+ cells predominated in the MLN and spleen, the number of lung CD8+ IL-10/eGFP+ cells was more than two folds higher than the number of lung CD4+ IL-10/eGFP+ cells (Fig. 4f and Supplementary Fig. 7 online). Taken together, these data demonstrate that the production of IL-10 in the influenza-infected lungs is exclusively restricted to the infiltrating effector T cells with CD8+ Te predominating over the CD4+ Te as contributors to the Te-derived IL-10.
In vivo blockade of IL-10 signaling during infection leads to lethal pulmonary inflammation
To test the function of IL-10 during influenza infection, we blocked IL-10 signaling in vivo by administration of a blocking IL-10R-specific mAb to mice undergoing minimally lethal infection with influenza. We found that blockade of IL-10 signaling resulted in increased and accelerated mortality (Fig. 5a). Of note, blockade of IL-10 action did not alter the titer of infectious virus in the lungs or virus clearance (Fig. 5b and Supplementary Fig. 8 online). We next examined whether the increased mortality observed with IL-10R-specific mAb administration was due to enhanced pulmonary inflammation. To explore this mechanism, we first surveyed the BALF of infected mice, treated with blocking antibody or isotype control antibody, for the production of a panel of inflammatory mediators. We found that a number of these mediators were elevated relative to infected controls in the BALF of IL-10R-specific mAb treated mice (Fig. 6a and Supplementary Fig. 9 online).
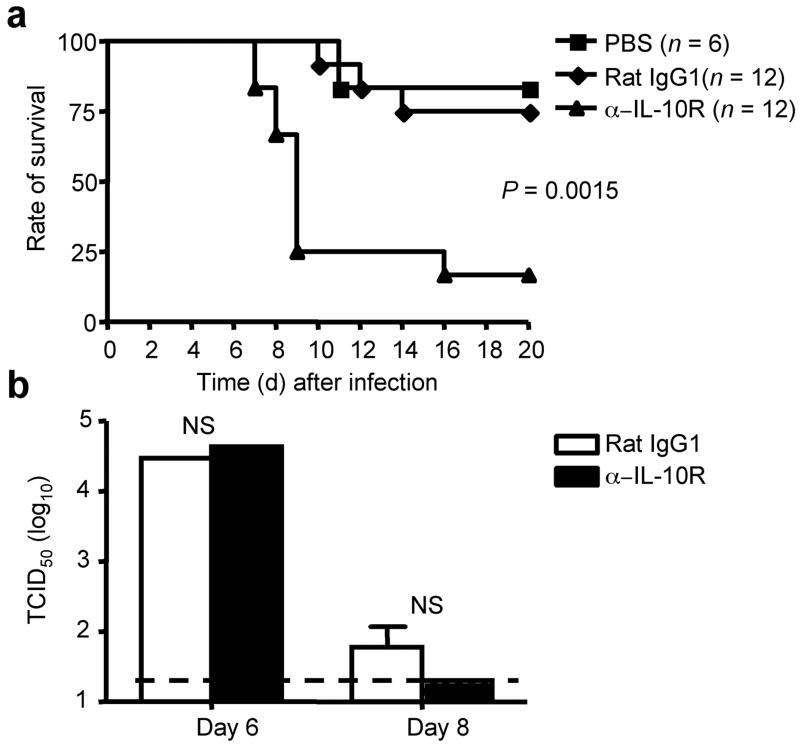
Figure 5. IL-10R blockade in vivo results in increased mortality and accelerated death during influenza infection.
(a) BALB/c mice were infected with influenza and treated with PBS, Rat IgG1 control mAb (Rat IgG1) or IL-10R-specific mAb (α-IL-10R). The survival of the mice was monitored daily. P value was determined by the Log-Rank survival test. (b) BALB/c mice were infected with influenza and treated with Rat IgG1 control mAb (Rat IgG1) or IL-10R-specific mAb (α-IL-10R). At day 6 and 8 p.i., airway (BALF) virus titers were assessed. P value was determined by unpaired two-tailed Student t test.
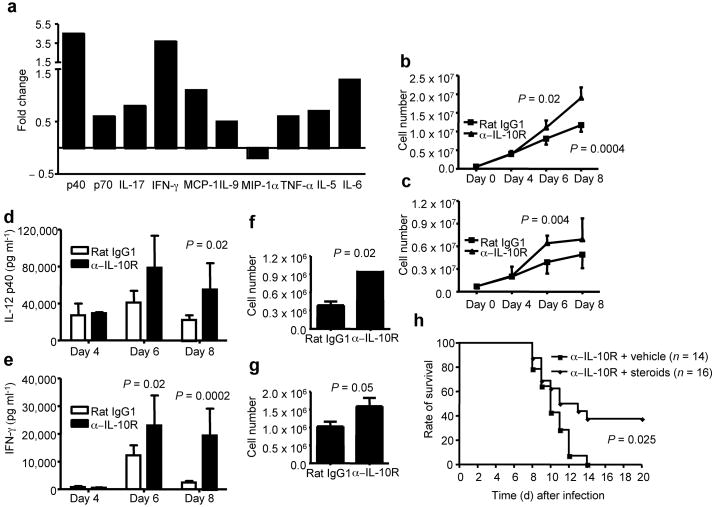
Figure 6. IL-10R blockade leads to lethal pulmonary inflammation during influenza infection.
(a) BALB/c mice were infected with influenza and treated with IL-10R-specific mAb (α-IL-10R) or Rat IgG1 control mAb (Rat IgG1). At day 8 p.i., cytokines in the BALF were determined by Multi-plex cytokine array analysis. Shown are the fold changes of cytokines in the BALF of IL-10R mAb treated mice vs. those of cytokines in the BALF of Rat IgG1 treated mice. (b, c) BALB/c mice were infected with influenza and treated with IL-10R-specific mAb (α-IL-10R) or Rat IgG1 control mAb (Rat IgG1). At day 4, 6 and 8 p.i., lung inflammatory monocytic cells (b) and neutrophils (c) were determined by flow cytometry. P value was determined by unpaired two-tailed Student t test. (d, e) BALB/c mice were infected with influenza and treated with IL-10R-specific mAb (α-IL-10R) or Rat IgG1 control mAb (Rat IgG1). At day 4, 6 and 8 p.i., IL-12 p40 (d) and IFN-γ (e) levels in the BALF were determined by ELISA. P value was determined by unpaired two-tailed Student t test. (f, g) BALB/c mice were infected with influenza and treated with IL-10R-specific mAb (α-IL-10R) or Rat IgG1 control mAb (Rat IgG1). At day 8 p.i., lung cells were restimulated with flu infected BMDC and the numbers of IFN-γ+ CD4+ (f) and IFN-γ+ CD8+ (g) T cells were quantified by ICS. P value was determined by unpaired two-tailed Student t test. (h) BALB/c mice were infected with influenza and treated with IL-10R-specific mAb plus vehicle control (α-IL-10R + vechicle) or IL-10R-specific mAb plus corticosterone (α-IL-10R + steroids). The survival of the mice was monitored daily. P value was determined by the Log-Rank survival test.
We also surveyed potential cellular targets of IL-10 action in the lungs and found (Supplementary Fig. 10 online) that the IL-10 receptor was preferentially expressed on inflammatory monocytic cells of the dendritic cell and macrophage lineages. Furthermore, we observed a substantially increased numbers of the inflammatory monocytic cells infiltrating to the lungs after IL-10R blockade in vivo (Fig. 6b). IL-10R blockade also had a modest effect on neutrophil accumulation in the lungs (Fig. 6c). The marked increase in inflammatory monocytic cells was associated with increased production in BALF of IL-12p40, a cytokine that is primarily produced by monocytic cells (Fig. 6d). IL-10R blockade also resulted in an increase in the level of IFN-γ released into BALF (Fig. 6e). This increased production of IFN-γ observed with IL-10R blockade was associated with a modest increase in the number of virus-specific Te (Fig. 6f, g). Interestingly, high-dose influenza virus infection also leaded to a massive influx of monocytic cells and neutrophils, associated with a decrease in IL-10 production (Supplementary Fig. 11 online).
The above results strongly suggested that blocking of Te--mediated IL-10 signaling results in increased inflammation and potentially lethal injury during acute influenza infection, without impacting virus titer or clearance. Therefore, we reasoned that suppression of the exaggerated inflammatory response produced by IL-10R blockade during influenza infection may prevent the development of lethal pulmonary injury and death. To evaluate this, we examined the effect of corticosteroid administration on the survival of infected mice undergoing IL-10R blockade. We found that corticosteroid administration partially protected infected mice from lethal injury and death (Fig. 6h). Importantly, steroid administration at a time immediately prior to the influx of Te into the infected lungs suppressed inflammation as monitored by the recruitment of monocytic cells to the lungs and IL-12 p40 levels in BALF, but had minor effects on lung virus titers and virus clearance (Supplementary Fig. 12 online).
DISCUSSION
In this report we analyzed the production and function of the regulatory/anti-inflammatory cytokine, IL-10, in the respiratory tract during acute experimental influenza infection. We found that, in contrast to the persistent low-level of IL-10 produced during chronic viral infection 6–8, acute influenza infection induces rapid and transient high-level production of IL-10 in the infected respiratory tract coincident with onset of the adaptive immune response. The source of this IL-10 is anti-viral CD8+ and CD4+ Te themselves, with CD8+ Te likely serving a greater contributor to the total Te-derived IL-10 in the lungs than CD4+ Te. Importantly, we demonstrate that this Te-derived IL-10 plays a crucial role in regulating the development of lung inflammation and lethal injury. Thus, our findings reveal a novel regulatory role for anti-viral Te in controlling excess inflammation and associated immune-mediated pathology during acute respiratory virus infection.
While not definitive, multiple lines of evidence reported here suggest that anti-viral CD8+ Te are a major source of the IL-10 produced in the influenza infected lungs. Several earlier reports 18,19 as well as more recent data 20,21 indicated that activated CD8+ T cells were capable of secreting IL-10. However, it was not established whether IL-10, under these circumstances, was produced by conventional CD8+ Te or a “ regulatory” CD8+ T cell subset 22. Indeed, IL-10 producing CD8+ T-cells were also detected (by ELISPOT) in lymph nodes draining the lung 23 and IL-10 transcripts were detected in a fraction of total CD8+ T-cells isolated from infected lungs 24 in experimental influenza infection. However, in these instances, it was not determined if virus-specific CD8+ T cells produce IL-10 protein in vivo in the infected lungs. To our knowledge, this is the first report demonstrating high-level production of IL-10 by Te in acute virus infection, as well as the biologically important impact of such Te-derived IL-10 on the outcome of infection. We speculate such a mechanism may operate in a variety of acute virus infections where both intensive inflammation and strong Te responses are induced.
Consistent with the critical role of CD8+ Te in virus clearance, IL-10 producing CD8+ Te are highly enriched at the site of infection. Interestingly, we found that the induction of these IL-10 producing CD8+ Te in the lung is dependent on the presence of CD4+ T cells (Fig. 4c). Importantly, in agreement with published data 25,26, CD4+ T cells are needed neither for the accumulation of CD8+ Te nor for the production of IFN-γ and TNF-α by these cells in vivo during acute influenza infection (Fig. 4c and data not shown). Thus, these data raises the possibility that “help” from CD4+ T cells can shape the quality of CD8+ T cell responses (e.g. regulatory cytokine production) during acute virus infection.
Blockade of IL-10 produced by CD8+ and CD4+ Te resulted in enhanced lung inflammation, elevated expression of multiple cytokines and chemokines in the infected lungs and increased mortality with importantly no effect on virus titer or clearance. IL-10 is recognized as a regulatory (anti-inflammatory) cytokine. This cytokine can act on multiple cell types to regulate immune and inflammatory responses 4. Inflammatory cells of the myloid lineage particularly the monocytic lineage (e.g., inflammatory lung dendritic cells and macrophages) represent attractive and, likely, primary cellular targets for IL-10 in infected lungs. As we (Supplementary Fig. 10 online) and others 27 demonstrate, this cell lineage expresses IL-10R, and these cells increase in number in the infected lungs following IL-10R blockade. In this connection it is noteworthy that a subset of monocytic cells recruited into the influenza infected lungs has been implicated in the development of increased inflammation and associated immunopathology during experimental influenza infection 28. Although the mechanism of action of this Te-derived IL-10 has yet to be fully elucidated, IL-10 mediated inhibition of recruitment, activation and/or pro-inflammatory cytokine and chemokine production by these inflammatory monocytic cells may be critical for regulating excess inflammation during the host response to influenza infection 4. Such an effect of IL-10 could account for the aforementioned increase in cytokine/chemokine production as well as the increase in inflammatory cell accumulation in the infected lungs following IL-10R blockade. An associated increase in the activation state of the infiltrating inflammatory monocytic cells could also result in increased stimulation of recruited virus-specific Te and the elevated levels of IFN-γ detected in the infected lungs following blockade. IL-10 may also act directly on the Te themselves in an autocrine fashion to dampen the response of the Te to viral antigen. A more complete understanding of the mechanism(s) of control inflammation by IL-10 in this model will ultimately require the characterization of the cell types expressing IL-10R in the inflamed lungs and the response of these cell types to IL-10.
It is particularly noteworthy that blocking IL-10 activity during infection results in the overproduction of several proinflammatory mediators (e.g. TNF-α, IL-6 and MCP-1) that had been implicated in the “cytokine storm” observed in infections with the high pathogenic H5N1 avian and the “1918 Spanish” influenza viruses 1–3. These highly virulent influenza strains could produce exaggerated lung inflammation and lethal illness not only by enhancing pro-inflammatory responses 1–3, but also by suppressing the production of anti-inflammatory cytokines such as IL-10 by innate and, perhaps more importantly, by adaptive immune effector cells. In this connection, it is further noteworthy that we saw that high dose (lethal) influenza infection led to a disproportionate decrease in the production of IL-10 compared to IFN-γ in infected lungs. This finding raises the possibility that infection under conditions where virus titers reach high levels in the lungs early in infection, such as is observed with highly pathogenic influenza strains 29,30 or with high dose infection with conventional strains 31, may selectively suppress IL-10 production by Te. We also observed that corticosteroid administration following IL-10R blockade partially reversed the effect of IL-10 blockade. This is not surprising given the inhibitory effect of corticosteroids on transcription of a large number of proinflammatory cytokines and chemokines as well as of adhesion molecules and inflammatory enzymes 32. This result reinforces the view that IL-10 produced by Te acts primarily to control the extent of inflammation induced by the adaptive (and innate) immune response during infection. Although corticosteroids can target many different cell types 32, the major impact of corticosteroid treatment in this model of infection and IL-10 blockade was suppression of the expression of the inflammatory monocytic lineage cell product, IL-12 p40, again implicating a possible role of this cell lineage in the development of pulmonary inflammation and injury during infection.
In summary, our findings here reveal a previously unrecognized function of Te, in particular CD8+ Te, in providing an essential regulatory function: control of excessive inflammation and associated tissue injury during acute viral infection. Understanding the mechanism of the expression of IL-10 by CD8+ Te and the function of this Te-derived anti-inflammatory cytokine may provide insight into the pathogenesis of infection with highly virulent strains of influenza as well as provide a framework for the design of more effective treatment modalities against these lethal infections.
METHODS
Mice and infection
WT BALB/c and C57/B6 mice were purchased from Taconic Farms. Rag1−/− mice were purchased from Jackson labs. IL-10-IRES-eGFP reporter mice (Vert-X) were generated by insertion of a floxed neomycin-IRES-eGFP cassette between the endogenous stop site and the polyA site of Il10, then neomycin resistance marker was excised by breeding with Zp3-Cre mice and successful Cre-deletion was confirmed by Southern blot analysis 33. Animal care and experiments were performed in accordance with institutional and US National Institutes of Health guidelines and were approved by the Animal Care and Use Committee of the University of Virginia. Infectious stocks of mouse adapted influenza virus A/PR/8/34 (H1N1) were prepared as previously described 16. 12–15 week old Balb/c mice were infected with 500 egg infectious units (EIU) dose of PR/8 in serum free Iscove’s media intranasally following anesthesia with halothane.
Quantitative RT-PCR
Lung single cell suspensions were prepared as previously described 16. To measure cytokine expression, RNA from MACS-purified lung CD3+ CD3−, CD8+ or CD4+ cells was isolated with RNeasy kit (Qiagen)and treated with DNase I (Invitrogen). Oligo(dT) (Promega) andSuperscript II (Invitrogen) were used to synthesize first-strandcDNAs from equivalent amounts of RNA from each sample. Real-timeRT-PCR was performed in a 7000 Real-Time PCR System (AppliedBiosystems) with SYBR Green PCR Master Mix (Applied Biosystems). Data were generated with the comparative threshold cycle (DeltaCT) method by normalizing to hypoxanthine phosphoribosyltransferase (HPRT). Sequences of primers used in the studies are available on request.
T cell restimulation by BMDC
BMDC were generated as described 34. On day 6–7, BMDC were harvested and infected with influenza virus at approximated 100 EIU per cell for 5–6 h. Then BMDC were counted and mixed with total lung, MLN or spleen cells at a 1.5 to 1 ratio in the presence of Golgi-Stop (BD Biosciences, 1 μl ml−1) and hIL-2 (40 U ml−1) for additional 6h. The surface staining of cell surface markers, intracellular staining of cytokines, T-bet and Foxp-3 were performed as described before 35 or according to manufacturer manuals. For the measuring of granule exocytosis by CD107a, fluorescence labeled CD107a-specific mAb or isotype control mAb were added to the in vitro stimulation cultures and the surface accumulation of CD107a was measured by flow cytometry 36.
BrdU labeling in vivo
At day 6 p.i. with influenza, mice were injected with 3 mg BrdU (BD Biosciences) through i.v.. 2 h later, mice were sacrificed and lung cell suspensions were restimulated with flu infected BMDC. The staining of cytokines and BrdU was performed according to the manufacturer manuals.
T cell depletion in vivo
At day 3 p.i. with influenza, mice were injected with 200 μg CD8-specific mAb (Clone 2.43), 500 μg CD4-specific mAb (Clone GK1.5) or 200 μg CD8-specific mAb plus 500 μg CD4-specific mAb i.p. We confirmed the specific depletion of the mAb by flow cytometry (data not shown).
BALF cytokine determination
BALF was obtained by flushing the airway multiple times with a single use of 500 μl sterile PBS. Cells in BALF were spun down and supernatants were collected for ELISA analysis (BD Biosciences) or 23-plex cytokine array analysis (Bio-Rad) according to the manufacturer manuals.
In vivo ICS assay
Measurement of IL-10 and IFN-γ producing cells in vivo was based on a previously described protocol with modifications 37. Briefly, at day 6 p.i., mice were injected i.v. with 500 μl of a PBS solution containing 500 μg monensin (Sigma-Aldrich) 6 h before harvesting. Lung single cell suspensions were prepared in the presence of monensin. Cells were then fixed and permeablized and intracellular IL-10 and IFN-γ staining was as described 35.
IL-10R-specific mAb and corticosteroid administration in vivo
Blocking IL-10R-specific mAb (Clone 1B1.3A) and isotype control Rat IgG1 mAb were purchased from Bio-express. IL-10 signaling blockade in vivo were achieved by the injection of IL-10R blocking mAb on day 3 (i.p. 1mg, 500μl), day 4 (i.n. 0.15mg, 40μl) and day 6 (i.p. 1mg, 500μl). Corticosterone was purchased from Sigma-Aldrich. In vivo treatment with corticosterone and vechicle (10% ethanol in PBS) was achieved by i.p. injection of 1 mg corticosterone daily from day 5.5 to day 8.5.
Virus Titer
Lung viral titers were monitored via endpoint dilution assay and expressed as Tissue Culture Infections Dose 50 (TCID50). Briefly, MDCK cells were incubated with 10-fold dilutions of BALF or lung homogenate from influenza virus infected mice in serum free DMEM media. After 3 day incubation at 37°C in a humidified atmosphere of 5% CO2, supernatants were collected and mixed with an half volume of 0.5% chicken RBC, the agglutination pattern read, and the TCID50 values calculated. For virus tittering by real-time RT-PCR, RNA was extracted from lung homogenate using Trizol (Invitrogen). Reverse transcription and influenza polymerase (PA) gene quantification were performed as described before 38.
FACS analysis
All FACS Abs were purchased from BD Biosciences or eBioscience. The dilution of surface staining Abs was 1:200 and dilution of intracellular staining Abs was 1:100. After Ab staining, cells were acquired through a 6-color FACS-Canto system (BD Biosciences). Data were then analyzed by FlowJo software (Treestar). The different cell types were characterized accordingto their phenotype, as follows: neutrophils (Ly6g+ CD11bhigh Ly6c− SSCmed), monocytic cell lineage (Ly6g− CD11bhigh Ly6c+, NK cells (DX-5+ CD3−), B lymphocytes (B220+ FSClow SSClow), CD8+ Tlymphocytes (CD3+ CD8+), CD4+ T lymphocytes (CD3+ CD4+).
Statistical analysis
Data are mean ± s.d.. The Kaplan-Meier Log-Rank survival test, One-way Anova or Student’s t-test were used as indicated in the text. We considered all P values > 0.05 not to be significant.
Supplementary Material
Acknowledgments
We thank M. Hufford and T. Kim for critical comments and B. Small and S. Gill for excellent technical assistance. This work was supported by Natioinal Institutes of Health (AI15608, HL-33391 and AI37293 to T. J. B. and AI057992 to C.L.K.).
Footnotes
AUTHOR CONTRIBUTIONS
J.S. performed the experimental work and T.J.B. supervised the project. R.M. and C.L.K. provided a critical reagent for the study. The manuscript was written by J.S. and T.J.B. All authors approved the final manuscript.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Cheung CY, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–7. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 2.de Jong MD, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–7. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kobasa D, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–23. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 4.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 5.Couper KN, Blount DG, Riley EM. IL-10: The Master Regulator of Immunity to Infection. J Immunol. 2008;180:5771–7. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 6.Ejrnaes M, et al. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. 2006;203:2461–72. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks DG, et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–9. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humphreys IR, et al. Cytomegalovirus exploits IL-10-mediated immune regulation in the salivary glands. J Exp Med. 2007;204:1217–25. doi: 10.1084/jem.20062424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maynard CL, et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–41. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 10.O’Garra A, Vieira P. T(H)1 cells control themselves by producing interleukin-10. Nat Rev Immunol. 2007;7:425–8. doi: 10.1038/nri2097. [DOI] [PubMed] [Google Scholar]
- 11.Jankovic D, et al. Conventional T-bet(+)Foxp3(−) Th1 cells are the major source of host-protective regulatory IL-10 during intracellular protozoan infection. J Exp Med. 2007;204:273–83. doi: 10.1084/jem.20062175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson CF, Oukka M, Kuchroo VJ, Sacks D. CD4(+)CD25(−)Foxp3(−) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J Exp Med. 2007;204:285–97. doi: 10.1084/jem.20061886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groux H, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–42. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 14.Stock P, et al. Induction of T helper type 1-like regulatory cells that express Foxp3 and protect against airway hyper-reactivity. Nat Immunol. 2004;5:1149–56. doi: 10.1038/ni1122. [DOI] [PubMed] [Google Scholar]
- 15.Yoon H, Legge KL, Sung SS, Braciale TJ. Sequential activation of CD8+ T cells in the draining lymph nodes in response to pulmonary virus infection. J Immunol. 2007;179:391–9. doi: 10.4049/jimmunol.179.1.391. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence CW, Braciale TJ. Activation, differentiation, and migration of naive virus-specific CD8+ T cells during pulmonary influenza virus infection. J Immunol. 2004;173:1209–18. doi: 10.4049/jimmunol.173.2.1209. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence CW, Ream RM, Braciale TJ. Frequency, specificity, and sites of expansion of CD8+ T cells during primary pulmonary influenza virus infection. J Immunol. 2005;174:5332–40. doi: 10.4049/jimmunol.174.9.5332. [DOI] [PubMed] [Google Scholar]
- 18.Tanchot C, et al. Modifications of CD8+ T cell function during in vivo memory or tolerance induction. Immunity. 1998;8:581–90. doi: 10.1016/s1074-7613(00)80563-4. [DOI] [PubMed] [Google Scholar]
- 19.Endharti AT, et al. Cutting edge: CD8+CD122+ regulatory T cells produce IL- 10 to suppress IFN-gamma production and proliferation of CD8+ T cells. J Immunol. 2005;175:7093–7. doi: 10.4049/jimmunol.175.11.7093. [DOI] [PubMed] [Google Scholar]
- 20.Noble A, Giorgini A, Leggat JA. Cytokine-induced IL-10-secreting CD8 T cells represent a phenotypically distinct suppressor T-cell lineage. Blood. 2006;107:4475–83. doi: 10.1182/blood-2005-10-3994. [DOI] [PubMed] [Google Scholar]
- 21.Stumhofer JS, et al. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat Immunol. 2007;8:1363–71. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 22.Smith TR, Kumar V. Revival of CD8+ Treg-mediated suppression. Trends Immunol. 2008;29:337–42. doi: 10.1016/j.it.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Sarawar SR, Doherty PC. Concurrent production of interleukin-2, interleukin-10, and gamma interferon in the regional lymph nodes of mice with influenza pneumonia. J Virol. 1994;68:3112–9. doi: 10.1128/jvi.68.5.3112-3119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyle AG, Buttigieg K, Groves P, Johnson BJ, Kelso A. The activated type 1-polarized CD8(+) T cell population isolated from an effector site contains cells with flexible cytokine profiles. J Exp Med. 1999;190:1081–92. doi: 10.1084/jem.190.8.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belz GT, Wodarz D, Diaz G, Nowak MA, Doherty PC. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J Virol. 2002;76:12388–93. doi: 10.1128/JVI.76.23.12388-12393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shedlock DJ, et al. Role of CD4 T cell help and costimulation in CD8 T cell responses during Listeria monocytogenes infection. J Immunol. 2003;170:2053–63. doi: 10.4049/jimmunol.170.4.2053. [DOI] [PubMed] [Google Scholar]
- 27.Donnelly RP, Dickensheets H, Finbloom DS. The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J Interferon Cytokine Res. 1999;19:563–73. doi: 10.1089/107999099313695. [DOI] [PubMed] [Google Scholar]
- 28.Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol. 2008;180:2562–72. doi: 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- 29.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–2. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 30.Tumpey TM, et al. Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J Virol. 2005;79:14933–44. doi: 10.1128/JVI.79.23.14933-14944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legge KL, Braciale TJ. Lymph node dendritic cells control CD8+ T cell responses through regulated FasL expression. Immunity. 2005;23:649–59. doi: 10.1016/j.immuni.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Barnes PJ, Adcock IM. How do corticosteroids work in asthma? Ann Intern Med. 2003;139:359–70. doi: 10.7326/0003-4819-139-5_part_1-200309020-00012. [DOI] [PubMed] [Google Scholar]
- 33.Madan R, et al. Non-redundant roles for B cell-derived IL-10 in immune counter- regulation. doi: 10.4049/jimmunol.0900185. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lutz MB, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 35.Sun J, Pearce EJ. Suppression of early IL-4 production underlies the failure of CD4 T cells activated by TLR-stimulated dendritic cells to differentiate into Th2 cells. J Immunol. 2007;178:1635–44. doi: 10.4049/jimmunol.178.3.1635. [DOI] [PubMed] [Google Scholar]
- 36.Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Liu F, Whitton JL. Cutting edge: re-evaluating the in vivo cytokine responses of CD8+ T cells during primary and secondary viral infections. J Immunol. 2005;174:5936–40. doi: 10.4049/jimmunol.174.10.5936. [DOI] [PubMed] [Google Scholar]
- 38.Jelley-Gibbs DM, et al. Persistent depots of influenza antigen fail to induce a cytotoxic CD8 T cell response. J Immunol. 2007;178:7563–70. doi: 10.4049/jimmunol.178.12.7563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.