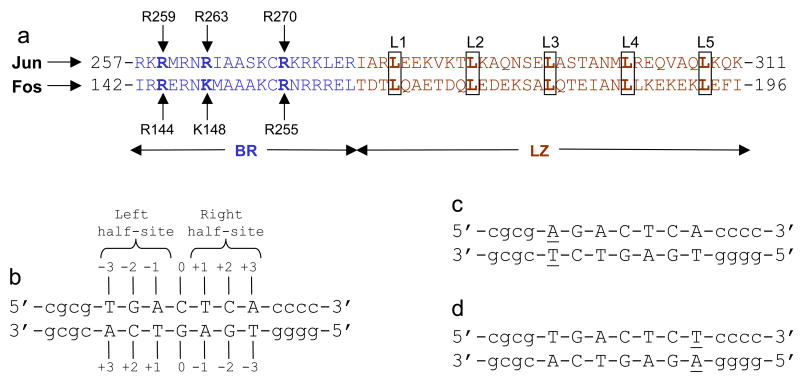
Figure 1.
Protein and DNA sequences. (a) Subdivision of bZIP domain into its respective N-terminal basic region (BR) and the C-terminal leucine zipper (LZ) for Jun and Fos transcription factors. The BR and LZ subdomains are colored blue and brown, respectively. The five signature leucines (L1-L5) characteristic of LZ subdomains are boxed and bold faced. The basic residues within the BR subdomains that hydrogen bond with specific DNA bases are labeled by vertical arrows. (b) Nucleotide sequence of 15-mer dsDNA oligo containing the TGACTCA motif. The TGACTCA motif is capitalized whilst the flanking nucleotides are shown in small letters. The numbering of various nucleotides relative to the central C/G base pair (assumed to be at zero position) in both strands is indicated. The TGA and TCA half-sites within this motif are also marked. (c) Nucleotide sequence of dsDNA oligo containing the T→A mutation at −3 position and herein referred to as the A-3 oligo. The variant nucleotides in both strands are underlined. (d) Nucleotide sequence of dsDNA oligo containing the A→T mutation at +3 position and herein referred to as the T+3 oligo. The variant nucleotides in both strands are underlined. Note that the A-3 and T+3 oligos shown in (c) and (d) are examples of a pair of symmetrically related dsDNA oligos in that they contain identical half-sites in opposite directions— these half-sites are indistinguishable upon the rotation of the variant motif by 180° in the plane of the paper (two-fold symmetry).