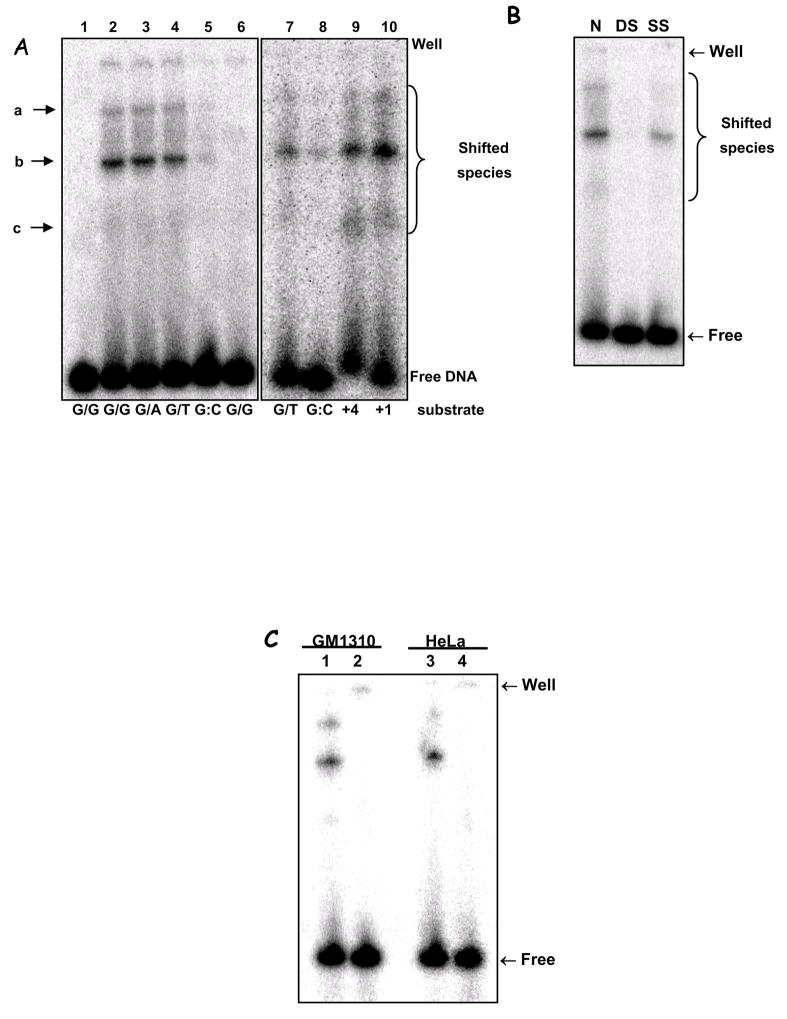
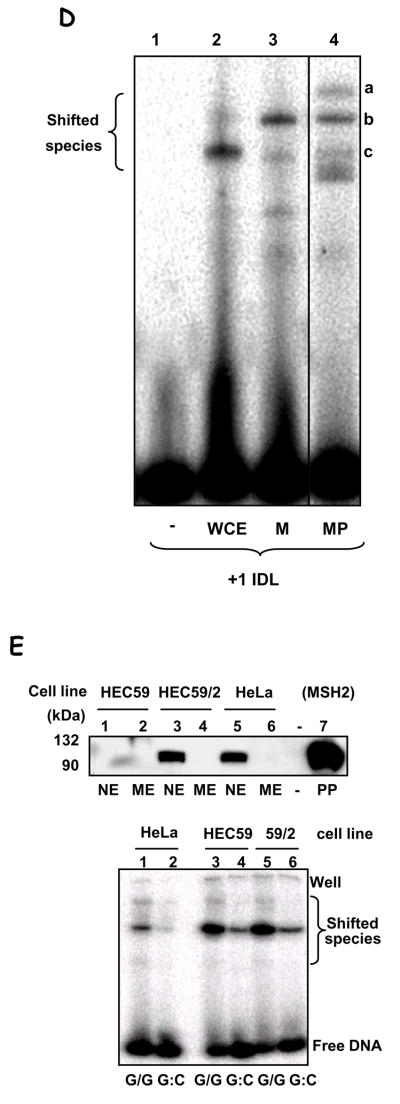
Figure 3.
A. EMSA of mismatch-containing substrate with HeLa mitochondrial extracts. Substrates: dG/dG (L1, 2 & 6), dG/dA (L3), dG/T (L4 & 7), dG:dC (L5 & 8), 4 bp- and 1 bp-insertion (+4 & +1, L9 & 10). L1 shows substrate alone; L6, heat-inactivated lysate (‘▲’, 95°C, 5 min). Electrophoretic origin: ‘well’, shifted species, and free substrate are indicated. Binding data is given in the text; N=3, values are mean ±SEM. B. Competition assays using unspecific competitor DNA were carried out essentially as in (A), with labelled dG/dG substrate, without (N) or with the addition of 20 ng of each poly[dI-dC]•[dI-dC] (DS), poly[dI-dC] (SS). C. EMSA was performed using dG/dG substrate, as in (A), heated at 95°C for 10 min (lanes 2 and 4) or not (lanes 1 and 3), and analyzed as above. Mitochondrial extracts from GM1310 (lanes 1 and 2) or HeLa cells (lanes 3 and 4) were analyzed. D. EMSA on +1IDL substrate with HeLa whole cell extract (WCE, lane 2), mitochondrial extract (M, lane 3), or mitoplast extracts (MP, lane 4). Lane 1 is substrate alone. E. Top panel: Western blot with hMSH2 antibody of nuclear (NE) and mitochondrial extracts (ME) from HeLa, HEC59 (msh2-deficient) and HEC59/2–5 (HEC59 complemented with chromosomes 2 and 4); purified hMutSα protein (L7, as MSH2/MSH6) is included as a positive control for MHS2 as indicated. Sizes are marked. Bottom panel: EMSA with mitochondrial extracts from HeLa (lanes 1 & 2), HEC59 (lanes 3 & 4) and HEC59/2–4 (lanes 5 & 6) cells on dG/dG (lanes 1, 3 & 5) mismatched or control dG:dC (lanes 2, 4 & 6) substrates.