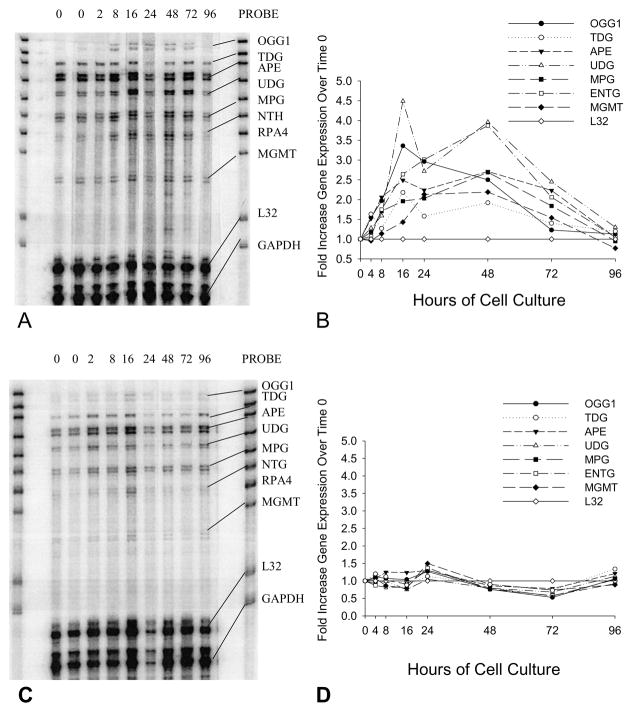
Fig. 1.
RPA results showing changes in the expression of seven base excision repair genes in MCF-10A cells treated without (A) or with (C) 0.1μM DOX for up to 96 hours. Results are representative of multiple experiments. Both cultures were started with cells synchronized in G0/G1 that did not achieve confluence. The PROBE lane shows the bands for each of the probes used in the assay. The probe lengths are greater than the “protected” fragment lengths because probes containing flanking sequences derived from the plasmid do not hybridize with target mRNAs. RP4 (subunit of RPA holoenzyme) expression was not detected in MCF-10A cells under the described experimental conditions. All gene expression values were normalized to the expression of L32 at the 0-hour point and fold increases/decreases over the expression of each gene at time zero were calculated for 2, 8, 16, 24, 48, 72, and 96 hours. (B) Quantification of selected DNA repair gene expression in MCF-10A cells cultured for 2, 8, 16, 24, 48, 72, and 96 hours following release from synchronization by addition of fresh medium. In these control cells, the expression of certain genes (mean ± SD; n=3) increased significantly following release from G1 block, including: OGG1 (2.5 ± 0.1 fold at 48 hours, P<0.01); TDG (2.2 ± 0.4 fold at 16 hours, P<0.05); APE (2.5 ± 0.3 fold at 16 hours, P<0.01); and UDG (4.0 ± 0.7 fold at 48 hours, P<0.02). (D) Quantification of the expression of selected DNA repair genes in MCF-10A cells cultured in the presence of 0.1μM DOX in fresh medium for 2, 8, 16, 24, 48, 72, and 96 hours.