Abstract
Camptothecins kill mammalian cells by stabilizing topoisomerase I-DNA strand passing intermediates that are converted to lethal double strand DNA breaks in DNA replication fork collisions. Camptothecin-stabilized topoisomerase I-DNA cleavage intermediates in mammalian cells are uniquely modified by ubiquitin-family proteins. The structure, composition and function of these ubiquitin-family modifications are poorly understood. We have used capillary liquid chromatography-nanospray tandem mass spectrometry to analyze the endogenous ubiquitin-family modifications of topoisomerase I purified from camptothecin-stabilized topoisomerase I-DNA cleavage complexes in human breast cancer cells. Peptides shared by SUMO-2 and SUMO-3 were abundant, and a peptide unique to SUMO-2 was identified. Ubiquitin was also identified in these complexes. No SUMO-1 peptide was detected in human topoisomerase I-DNA cleavage complexes. Identical experiments with purified SUMO paralogs showed that SUMO-1 was well digested by our protocol and that fragments were easily analyzed by LC-MS/MS. Spiking experiments with purified SUMO paralogs determined that we could detect as little as 0.5 SUMO-1 residue per topoisomerase I molecule. These results indicate that SUMO-1 is below this detection level and that SUMO-2, or a mixture of SUMO-2 and SUMO-3 predominates. SUMO-1 capping seems unlikely to be limiting the growth of SUMO-2/3 chains formed on camptothecin stabilized topoisomerase I-DNA cleavage complexes.
Ubiquitin-family modifiers are small proteins that are covalently attached to other proteins to regulate proteolysis, transport, localization, and other functions (1,2). SUMO-1 was the first member of the SUMO1 (Small Ubiquitin-like MOdifier) family (3). Two other SUMO paralogs, SUMO-2 (Swiss-Prot P61956) and SUMO-3 (Swiss-Prot P55854) have only about 43–42% homology to SUMO-1 but are about 96% identical to one another. With such a high degree of homology, SUMO-2 and SUMO-3 may be functionally equivalent, and are often designated SUMO-2/3. Recently, mice lacking SUMO-1 have been shown to be viable, with SUMO-2/3 being able to functionally substitute for it with only apparently minor effects (4). The much smaller sequence difference between SUMO-2 and SUMO-3 suggests that they may serve much the same functions in cells. However, the conservation of these two distinct paralogs in vertebrates hints at some selection pressure.
Ubiquitination and sumoylation of proteins are accomplished by similar systems of specialized enzymes (5). Sumoylation starts with activation by formation of a thioester linkage between SUMO and the E1 (SUMO-activating enzyme) enzyme, which then transfers SUMO to the conjugating enzyme, Ubc9, an E2 protein (SUMO carrier protein), in a transesterification reaction. Ubc9 then transfers the SUMO residue to a target protein, usually at ΨKX(E/D) (the sumoylation consensus sequence, where Ψ is a large hydrophobic amino acid, × is any amino acid, and the 4th amino acid is either E or D). Ubc9 does not discriminate between SUMO paralogs, and can sumoylate most target proteins in vitro, however, the majority of in vivo sumoylation reactions requires an E3 protein which guides Ubc9 to specific protein targets (5). Sumoylation and de-sumoylation are highly dynamic. Known functions for sumoylation include antagonism of ubiquitin dependent degradation, nuclear-cytoplasmic trafficking, targeting to subnuclear structures, regulation of transcription, mitochondrial fission, cell division, DNA damage repair, genetic instability, and assembly of protein complexes (5).
Sumoylation is precisely controlled in vivo. Intracellular distributions, mobility, and protein modification patterns by SUMO-1 and SUMO-2/3 are very different (6–9). Heat shock and oxidative stress cause depletion of the free SUMO-2/3 pool and SUMO-2/3 conjugation to high molecular weight proteins while having no effect on the distribution of SUMO-1, which remains on RanGap1 during these stresses. Such precise targeting of different SUMO paralogs indicates levels of regulation that remain incompletely understood (8,10). A mechanism for selective targeting of SUMO-2/3 paralogs to specific proteins by their preferential binding to SUMO Interaction Motifs (SIMs) has been reported (11,12). However, the SIM binding regions of SUMO-2 and SUMO-3 are identical (13). Since sumoylation can promote protein interactions and localization, the small differences in SUMO-2 and SUMO-3, localized to the N-terminal region, may favor binding of different proteins to a sumoylated target protein.
Topoisomerase I (topo I), an enzyme that removes superhelical stress generated by DNA replication and transcription, is the target of an important class of anticancer drugs, camptothecins (CPTs) (14). Like the parent drug, camptothecin (CPT), CPTs stabilize an intermediate in the topo I DNA strand passing reaction in which the topo I is covalently attached to the DNA at a strand break (15). CPTs are toxic to proliferating cells due to DNA replication fork collisions with the CPT-stabilized covalent topo I-DNA cleavage complexes (CptTop1cc) resulting in complex DNA lesions that include double strand DNA breaks, gaps, and ligations of parental DNA strands to nascent strands (16,17). When mammalian topo I trapped in CptTop1cc by CPT is released by nuclease digestion, it shows a laddered pattern of bands, extending to very high molecular weights (18), and attributed to numerous SUMO-1 additions to the topo I (19).
Many functions for the SUMO modifications of CptTopIcc have been reported, including nuclear export, nuclear localization, degradation, repair of complexes, and enhancement of DNA cleavage (18–24). The possible role of sumoylation in degradation of CptTop1cc (20,21) is interesting since the ubiquitin and sumoylation pathways interact, or crosstalk, in the cases of PCNA and IκBα (25). Sumoylation of CptTop1cc has also been suggested to be involved in delocalization of topo I from the nucleolus following inhibition of transcription by RNA polymerase inhibitors or DNA damage (23,24). However, other studies do not support a role for sumoylation in topo I nucleolar delocalization (22,26–28), so the function of this sumoylation remains elusive. Defects in Ubc9 leading to alterations in global sumoylation increase sensitivity to topo I-targeting drugs, but also increase sensitivity to a wide range of other genotoxic treatments, underscoring the complexity of sumoylation and its relation to DNA damage (29).
Although the laddering of CptTop1cc is thought to be due to sumoylation, the topo I in CptTop1cc is also modified by ubiquitin and degraded by the proteasome pathway (18). Exposure to CPTs causes down regulation of topo I (18,30,31), and this down-regulation is an important determinant of cancer cell sensitivity to CPTs (20). Although the topo I released from CptTop1cc distributes in a ladder pattern on gel electrophoresis, Western blotting of these complexes for ubiquitin shows a distinctly different pattern, typically a high molecular weight smear, that does not match the laddered distribution of the bulk of topo I from CptTop1cc (20,22). This suggests that ubiquitin and SUMO may occur on separate subpopulations of topo I in CptTop1cc. Although topo I contains 18 candidate sumoylation sites, mutational inactivation of candidate sites has indicated that K117 in the N-terminal region of topo-I is the only significant topo I sumoylation site (22,24). This, together with the high levels of sumoylation observed, strongly suggests branching SUMO chains. The composition and structure of SUMO chains formed on CptTop1cc in vivo is not known, but it was shown that in vitro conjugation of SUMO-1 to human topo I resulted in formation of polymeric SUMO-1 chains attached to K117 (32), supporting earlier evidence of SUMO-1 modification of topo I in CptTop1cc (19,22). Some studies have found that SUMO-1 cannot form homogeneous chains due to the absence of an internal sumoylation consensus sequence (33). However, in in vitro reactions SUMO-1 clearly can form homogeneous SUMO-1 chains (32,34–37) with the SUMO-1 residues forming chains by isopeptide bond formation to other SUMO-1 residues at amino acids K7, K16, and K17 in the SUMO-1 N-terminal region (36). It has been suggested that sumoylation is much more efficient in vitro than in intact living cells (38). Topors, a dual ubiquitin and SUMO ligase for p53, has been shown to greatly facilitate the formation of polymeric SUMO-1 chains on topoisomerase I in vitro and in vivo, resulting in the formation of extended SUMO-1 chains attached to topoisomerase I mainly at K117 and to a lesser extent at K103 and K153 (39).
Because of the unresolved issues concerning the composition and structure of ubiquitin-family modifications of topo I in CPT-treated cells, we have purified CptTop1cc from human cells expressing only endogenous SUMO paralogs, and studied the ubiquitin family modifications by mass spectrometry.
MATERIALS AND METHODS
Cell Culture and Camptothecin Treatment
MCF-7 (human breast adenocarcinoma) cells were grown to 80% confluence in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (Invitrogen) and penicillin/streptomycin in a humidified 5% CO2 atmosphere at 37°C. For CPT treatment, growth medium was replaced with serum-free medium with10 µM CPT (Calbiochem, San Diego, CA) for 30 min at 37°C.
Isolation of CPT-Stabilized Covalent Topoisomerase I-DNA Complexes
After drug treatment, medium was removed and the cells were lysed immediately by the addition of 800 µl of lysis solution (8 M guanidinium chloride, 0.2 N NaOH and 5% β-mercaptoethanol) per 10 cm plate. The lysates were collected in 50 ml polypropylene centrifuge tubes, sheared with vortexing (4 stainless steel nuts 3 mm in diameter per tube) for 2 min, followed by heating at 65°C for 15 minutes. The sheared lysates from four plates were applied to a CsCl (cesium chloride) step gradient prepared by the successive layering of 200 µl of CsCl solution A and 2 ml volumes of CsCl solutions B through D (A, 1.75g/ml; B, 1.62 g/ml; C, 1.42 g/ml; D, 1.32 g/ml) in a polyallomer tube (14 × 89 mm). The samples were centrifuged in a Beckman SW41 rotor at 30,000 rpm for 20 hr at 20°C. After centrifugation the samples were collected in 0.6 ml fractions. Spectrophotometric measurement of absorbance at 260 nm and 280 nm was used to calculate the amount of DNA in each fraction. The DNA containing fractions were combined, dialyzed against distilled water (4°C), and ethanol precipitated. The DNA pellet was transferred to a microcentrifuge tube, and rinsed three times with ice-cold 70% ethanol.
The DNA pellets were air-dried briefly, solubilized in 200 µl of nuclease buffer (50 mM Tris HCl pH 8.0 and 1 mM MgCl2), then incubated at 37°C for 6 hr, with two additions (80 units each) of Omnicleave nuclease (Epicentre Biotechnologies, Madison, WI). The sample volume was reduced to 50 µl, and the sample was extracted four times with 1 ml of ice-cold acetone with vortexing. The insoluble material was pelleted by centrifugation at 14,000 rpm for 15 min at 4 °C. The air dried pellet was resuspended by vortexing in 1 ml of 100% ice-cold ethanol, and centrifuged at 14,000 rpm for 15 min at 4°C (four times). The final protein pellet was air dried and resuspended in sterile distilled water. Protein content was determined using the Bio-Rad DC protein assay (Bio-Rad, Hercules, CA).
Mass spectrometry
Trypsin and Chymotrypsin Digestion of the protein
Purified CptTop1cc samples were analyzed after trypsin or chymotrypsin digestion at the Ohio State University Mass Spectrometry and Proteomics Facility. Several buffers were used for tryptic digestion: Buffer A: 25 mM ammonium bicarbonate, Buffer B: 25 mM ammonium bicarbonate buffer containing 6% acetonitrile (final concentration) and Buffer C: 25 mM ammonium bicarbonate containing 1 M urea (final concentration). The protein was diluted into 100 mM ammonium bicarbonate buffer (or 100 mM ammonium bicarbonate with 4 M urea), then 5 µl of a 5 mg/ml of DTT (dithiothreitol) in 100 mM ammonium bicarbonate buffer was added and incubated with the sample at 50°C for 15 min to reduce the cystines. The mixture was then brought to room temperature and 5 µl of a 15 mg/ml of iodoacetamide in 100 mM ammonium bicarbonate was added. The mixture was incubated in the dark for 15 min to alkylate the cysteines. Sequencing grade trypsin (Promega, Madison WI) was added to the mixture with an enzyme:substrate ratio of 1:25. Distilled water and/or acetonitrile were then added to the mixture to reach the desired final concentrations. Digestion was carried out at 37°C for 30 or 60 min and stopped by acidification. The digested fragments were analyzed by LC/MS/MS and the results were searched with MASCOT (Matrix Science, Boston, MA) for protein identification. Similar sequence coverage was obtained when the sample was digested with trypsin in buffers A-C. Chymotrypsin (Roche Diagnostic, Basel, Switzerland) digestion was performed using a similar approach except the final buffer composition was 25 mM ammonium bicarbonate and 5% acetonitrile.
Matrix-assisted laser desorption/ionization time-of- flight mass spectrometery (MALDI-TOF MS)
MALDI–TOF MS was performed on a Reflex III (Bruker Daltonics Inc, Billerica, MA) mass spectrometer. The instrument was calibrated with peptide standards with masses ranging from 500–2500 Da. The matrix, α-cyano-4-hydroxy cinnamic acid, was prepared as a saturated solution in 50% acetonitrile with 0.1% trifluoroacetic acid in water. Aliquots consisting of 5 µl of matrix and 1 µl of sample were thoroughly mixed, spotted on the target plate (1.0 µl) and allowed to dry before analysis. The instrument was operated in reflection, positive ion mode at an accelerating voltage of 28 kV. The N2 laser was operated at minimum threshold level required to generate signal and minimize dissociation. The instrument was calibrated with a mixture of 4 peptides (Bradykinin 1–5, Angiotensin I, Glu-Fib and ACTH 18–39). Data was acquired continuously using FlexControl software (Bruker Daltonics Inc, released in 2005) until suitable signal intensity was obtained. A peak list was generated using FlexAnalysis 2.4 (Bruker Daltonics Inc). The cut-off threshold for signal-to-noise ratio was set at 6 and resolution cut-off threshold was set at 5000. Peaks less than 800 Da were excluded from the peak list to avoid contaminations from the MALDI-TOF MS matrix. Only peaks with mass error less than 25 ppm, compared with theoretical mass, were considered as valid identification.
Nano-LC/MS/MS
Capillary-liquid chromatography-nanospray tandem mass spectrometry (Nano-LC/MS/MS) was performed on a Thermo Finnigan LTQ (linear quadrupole ion trap) mass spectrometer equipped with a nanospray source operated in positive ion mode. The LC system was an UltiMate™ Plus system from LC-Packings (Sunnyvale, CA) with a Famos autosampler and Switchos column switcher. Five microliters of each sample was first injected into the trapping column (LC-Packings), and washed with 50 mM acetic acid. The injector port was switched to inject, and the peptides were eluted off of the trap onto a 5 cm, 75 µm (inner diameter) ProteoPep II C18 reverse-phase column (New Objective, Inc. Woburn, MA) packed directly in the nanospray tip. Solvent A was 50 mM acetic acid in water and solvent B was acetonitrile. Peptides were eluted directly off the column into the LTQ system with a flow rate of 300 nl/min. The gradient was started and maintained at 2% B for the first 3 min, then B was increased to 50% in 27 min, and further increased to 80% in 15 min. B was kept at 80% for another 5 min, then returned to 2% in 0.1 min. The total run time was 65 minutes. The scan sequence of the mass spectrometer was programmed for a full scan and MS/MS scans of the 10 most abundant peaks in the spectrum. Dynamic exclusion was used to exclude multiple MS/MS of the same peptide after detecting it three times. Sequence information from the MS/MS data was processed using the Mascot 2.0 active Perl script with standard data processing parameters (first scan number, last scan number, number of different intermediate scans, minimum number of grouped scans, and minimum number of ions were set to 0, 0, 1, 0, and 8 respectively). Database searches were against the NCBInr 20060402 database using MASCOT 2.0 (Matrix Science). The mass accuracy of the precursor ions was set to 1.5 Da to accommodate accidental selection of the C13 ion, and the fragment mass accuracy was set to 0.5 Da. The number of missed cleavages permitted in the search was 2 for both tryptic and chymotryptic digests. Considered modifications (variable) were methionine oxidation, cysteine carbamidomethylation, phosphorylation, acetylation, and ubiquitin diglycine tags. An individual peptide was considered as a good match if it produced a probability-based MOWSE (MOlecular Weight SEarch) score (40) greater than 20 (P<0.05). The cut-off score for significant protein identification was set at 45 and the matched peptides were all manually verified.
Purified SUMO paralogs and Spiking Experiments
Purified human recombinant SUMO-1 (Boston Biochem UL-712, Cambridge, MA), SUMO-2 (Boston Biochem UL-752), SUMO-3 (Boston Biochem UL-762) were digested with trypsin and chymotrypsin for analysis by MALDI-TOF MS and LC/MS/MS as described for purified CptTop1cc. For spiking experiments, SUMO-1 was spiked into purified CptTop1cc with the molar ratios of 5:1, 1:1, 0.5:1, 0.2:1 and 0.1:1 (SUMO 1:Topo I). Briefly, 2.45, 0.49, 0.24, 0.12 and 0.049 µg of SUMO-1 was added into 4µg of purified CptTop1cc, respectively. The mixture was digested with trypsin or chymotrypsin using the digestion conditions described above for purified CptTop1cc. After 4 hours, the digestion was quenched by adding 0.1% trifluoroacetic acid. Samples were then concentrated until the final volume was 25µl, and 2.5µl of the sample was used for LC/MS/MS analysis. There is some confusion in the literature concerning the identity of SUMO-2 and SUMO-3. For our study, SUMO-2 is the protein identified as Swiss-Prot accession number P61956, with the amino acid sequence: MADEKPKEGV KTENNDHINL KVAGQDGSVV QFKIKRHTPL SKLMKAYCER QGLSMRQIRF RFDGQPINET DTPAQLEMED EDTIDVFQQQ TGG. SUMO-3 is Swiss-Prot accession number P55854, and differs from SUMO-2 (P61956) at the positions indicated by underlining in the SUMO-2 sequence above and the first 20 amino acids of the SUMO-3 sequence here: MSEEKPKEGV KTEN—DHINL. Many contributors to the field have designated Swiss-Prot P55854 as SUMO-2 and Swiss-Prot P61956 as SUMO-3 (9,33,41,42).
Electrophoresis and Western Blotting
Laemmli buffer (Bio-Rad) was added to each sample in amounts determined to equalize the protein concentration of each sample after the DNA digestion, acetone and ethanol washes. Samples were analyzed by one dimensional SDS-PAGE using a 7.5% separating gel and silver staining (Bio-Rad). Western blotting of CptTop1cc was done with rabbit anti-SUMO-2/3 (Zymed Laboratories, San Francisco, CA), goat anti SUMO-1 (Roche Applied Science, Indianapolis, IN), and human scleroderma anti-human topoisomerase I antibody (TopoGen, Port Orange, FL). Anti-ubiquitin antibody was a kind gift of Arthur Haas, and has been described (43). Chemiluminescent detection was done using Supersignal West Pico substrate (Pierce, Rockford, IL), x-ray film, and horseradish peroxidase conjugated secondary antibodies (Roche Applied Science, and Pierce). Acrylamide gradient gel electrophoresis was done using pre-formed 4–15% gels (BioRad).
RESULTS
Isolation of covalent topoisomerase I-DNA complexes
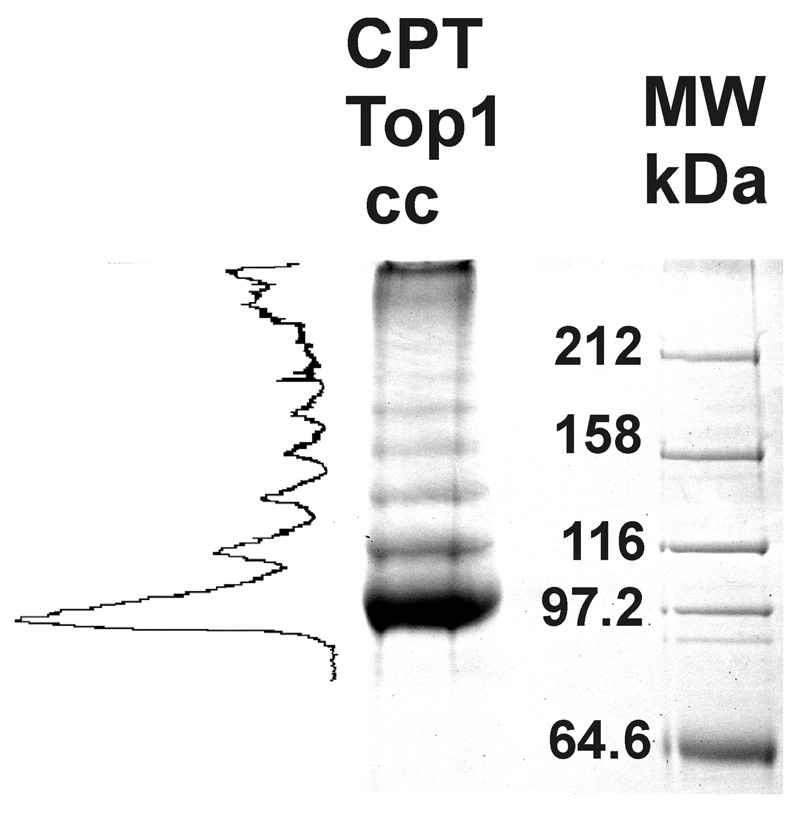
Proteins released by nuclease digestion of the purified DNA of CPT-treated cells showed a pronounced ladder pattern by Coomassie Brilliant Blue staining (Figure 1). We allowed the Omnicleave nuclease (about 26.6 kDa) to run off the gel. This ladder pattern has been seen by Western blotting of CptTop1cc released from DNA by shearing or nuclease digestion and analyzed by Western blotting in cell lysates (19), and consists of a major unmodified topo I band, followed by more slowly migrating sumoylated topo I bands which become more closely spaced at the higher levels of sumoylation until they are unresolved. The major band (band 1) had a molecular weight of about 104 kDa and seven bands of higher molecular weight were clearly visible (115 kDa, 137 kDa, 160 kDa, 190 kDa, 202 kDa, and 214 kDa.). Some of the band spacings correspond to the approximate molecular weights of sumo proteins (~11 kDa) and some to twice this value (or more). Free sumo proteins are known to migrate anomalously on SDS gels, at ~ 20 kDa, or twice their actual molecular weights (3). Anomalous migration on SDS gels is known for many proteins, including topoisomerase I. Anomalous migration may be due to retention of some peptide conformation or uneven SDS binding. The branching of SUMO chains may also contribute to anomalous migration, so that exact matches of the spacing to SUMO molecular weights should not be expected. The Coomassie stained CptTop1cc were analyzed by direct laser densitometry of the stained gel. Integration of the densitometry trace (Figure 1, left of the gel image) showed that band 1 (104 kDa) represents 44% of the Coomassie stained protein in the gel, and the more slowly migrating sumoylated forms represent 56%. These results indicate that our isolation of CptTop1cc results in the expected gel pattern reflecting extensive modification of the topo I by ubiquitin-family proteins. We believe this is the first quantitation of the ubiquitin family modifications of CptTop1cc by a biochemical method.
FIGURE 1.
Protein crosslinked to cellular DNA by camptothecin treatment of cells. Protein covalently bound to the DNA of CPT-treated post-confluent MCF-7 cells (4.5 × 107 cells) was isolated as described, separated by SDS PAGE, and stained with Coomassie Brilliant Blue. Global adjustments of brightness and contrast (Photoshop) were made for the digital image to compensate for the flattening effects of digital photography and show bands visible to the eye. A laser densitometric tracing (LKB UltroScan XL) of the stained gel is shown to the left of the gel image and molecular weight markers are shown to the right. Densitometry was done directly through the stained gel, not from the image.
To further characterize the isolated CptTop1cc we used acrylamide gradient gel electrophoresis (Supporting Figure 1S). Lanes loaded with only the Omnicleave nuclease used to release CptTop1cc from cellular DNA showed only a single band at a position corresponding to a molecular weight of ~27 kDa. Lanes loaded with purified CptTop1cc showed the ladder of topoisomerase I and higher molecular weight bands of sumoylated topoisomerase I. Control samples from cells that were treated with solvent only and processed identically to those from the CPT treated cells were free of protein bands, except for a single band of Omnicleave nuclease. The gradient gel showed faint smears below the main topoisomerase I bands. This was not detected in non-gradient gels such as that in Figure 1, or in topoisomerase I Western blotting (see below) of CptTop1cc. Since the faintly staining smear was not detected in the control lanes of the gradient gel (cells not treated with CPT), it is dependent on CPT treatment, and we conclude that it represents heterogeneous proteolytic fragments of CptTop1cc that were released from DNA by nuclease digestion. We feel that our alkaline GuHCl lysis with mercaptoethanol is very efficient at denaturation and inactivation of proteases, so this may represent proteolytic processing of CptTop1cc that occurs during CPT treatment of the cells.
Proteomic Analysis of Camptothecin-Stabilized Topoisomerase I-DNA Complexes
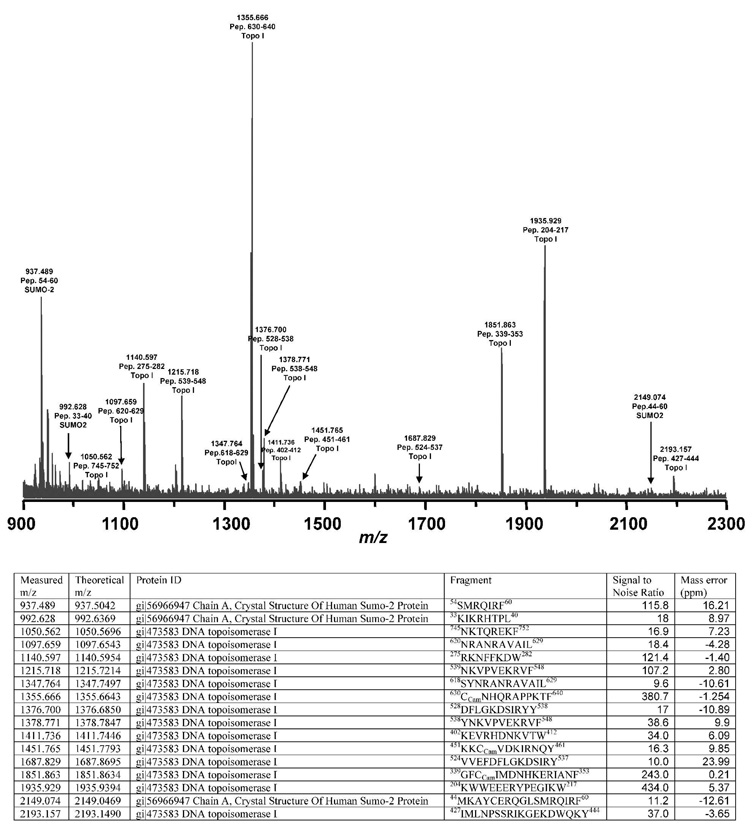
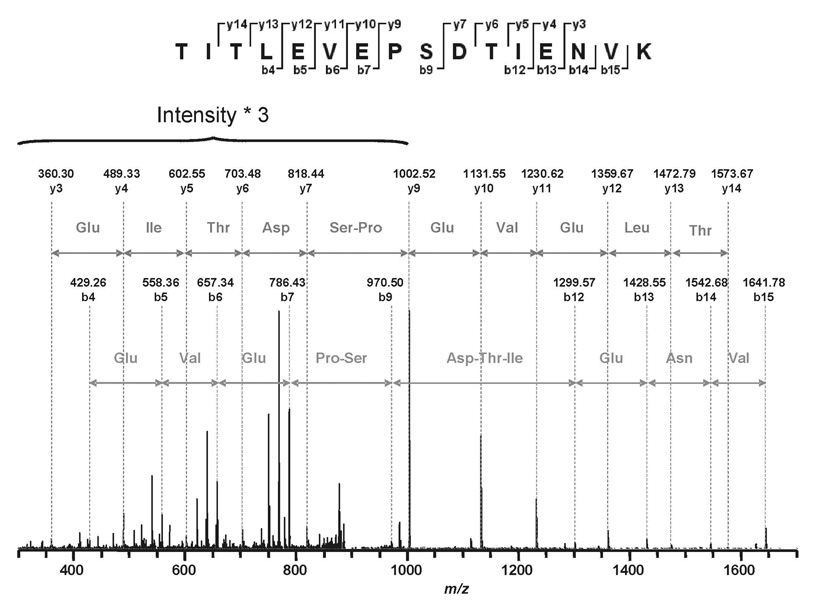
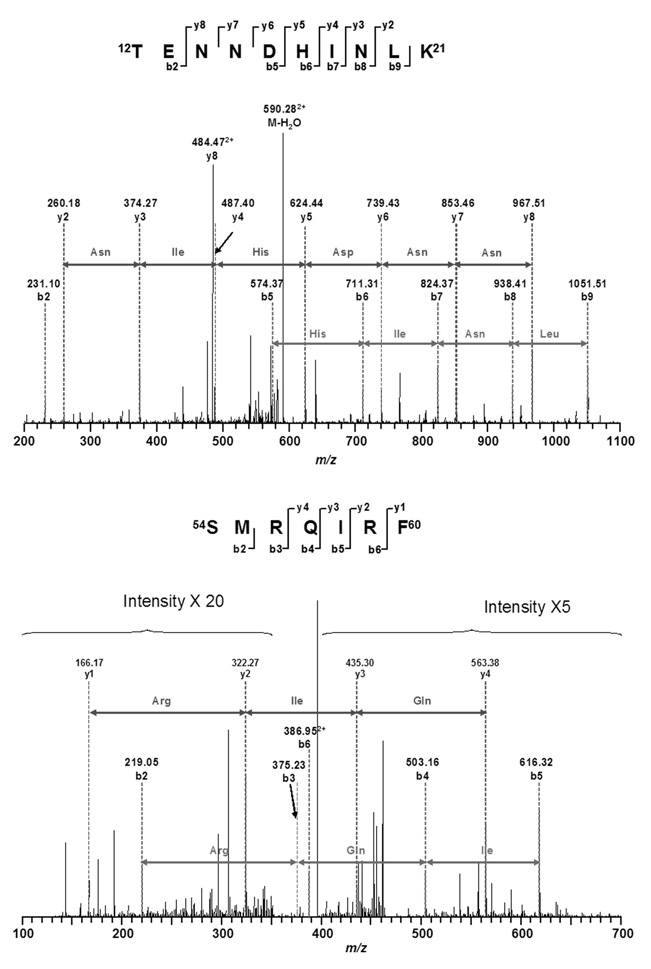
MALDI-TOF MS of the CptTop1cc samples identified human topo I peptides and SUMO-2/3 peptides as summarized in Figure 2. LC/MS/MS analysis of the CptTop1cc samples using trypsin and chymotrypsin digestions identified human topo I with 76.2% sequence coverage (Figure 3). SUMO-2 was identified with 67.7% sequence coverage, and ubiquitin was identified with 21.1% sequence coverage. No peptides of other human topoisomerases were detected and no SUMO-1 peptides were detected. One peptide at 894.802+ was observed and was sequentially identified as 12TITLEVEPSDTIENVK27 from ubiquitin (mass error 0.6727 Da, MASCOT score 56). The fragment ion sequence assignments of this ubiquitin peptide are shown in Figure 4 and Table 1S (Supporting Information). Many of the peptides of SUMO-2 are shared with SUMO-3. However, one tryptic fragment, 12TENNDHINLK21 at 599.58+2 (mass error 0.5742 Da, Mascot score 43), is unique to SUMO-2. The corresponding peptide TENDHINLK, unique to SUMO-3, was not detected. The fragment ion sequence assignments for 12TENNDHINLK21 and for a peptide common to SUMO-2 and SUMO-3 (54SMRQIRF60) are shown in Figure 5 and Table 2S (Supporting Information). No peptides of proteins other than topoisomerase I, ubiquitin, and SUMO-2/3 were detected. No phosphorylated or acetylated peptides were detected, and no peptides with masses expected for topoisomerase I or SUMO-1, SUMO-2, or SUMO-3 with diglycine tags (indicating ubiquitination at lysines) were detected.
FIGURE 2.
MALDI-TOF MS analysis of camptothecin-stabilized covalent human topoisomerase I-DNA complexes. A, MALDI-TOF mass spectrum. B, list of peptides identified. The SUMO-2 peptides identified by MALDI-TOF MS are shared with SUMO-3.
FIGURE 3.
LC/MS/MS Sequence coverage for camptothecin-stabilized covalent human topoisomerase I-DNA complexes. Amino acids observed in the tryptic digest, the chymotryptic digest, and both are indicated.
FIGURE 4.
Sequence assignments for a ubiquitin peptide observed in purified camptothecin-stabilized topoisomerase I-DNA complexes.
FIGURE 5.
Sequence assignments for the unique SUMO-2 peptide, TENNDHINLK and the SUMO-2/3 shared peptide SMRQIRF.
Proteomic Analysis of Camptothecin-Stabilized Topoisomerase I-DNA Complexes from heat shocked cells
Heat shock is known to prevent sumoylation of topo I in CptTop1cc (23,44) so that only a single band is obtained upon electrophoresis, rather than the sumoylation ladder typically seen. We have confirmed this (Fig 2S, Supporting Information), and analyzed the CptTop1cc isolated from heat shocked cells by LC/MS/MS. Only peptides from human topo I (29.5 % sequence coverage) were detected (Figure 2S, Supporting Information). No peptides matching ubiquitin family proteins were detected. Although CsCl density gradient centrifugation is well established as a way to cleanly separate cellular DNA from free proteins, this gel and LC/MS/MS result additionally demonstrate that the ubiquitin family proteins identified in CptTop1cc (Figure 2–Figure 5) are covalently attached to the topo I in CptTop1cc, and do not co-isolate with cellular DNA under the denaturing conditions of the alkaline CsCl gradient.
Purified SUMO Paralogs and SUMO Spiking Experiments
The absence of SUMO-1 in purified CptTop1cc was surprising, given the literature suggesting that these complexes are extensively modified by SUMO-1. To rule out the possibility that SUMO-1 does not digest well under the conditions used, or that the SUMO-1 peptides are present but not detected in the nanospray experiments, we carried out experiments with purified recombinant SUMO paralogs and with purified CptTop1cc spiked with purified SUMO-1. Experiments with recombinant SUMO-1 showed no problems with protease digestions, MALDI-TOF MS, or LC/MS/MS detection of SUMO-1 peptides, with 87% overall sequence coverage (Fig. 3S and Fig. 4S, and Table 3S, Supporting Information). Recombinant SUMO-2 gave 77% overall sequence coverage and recombinant SUMO-3 gave 52% sequence coverage (Figure 3S, Supporting Information). These results suggest that SUMO-1 digests at least as well as SUMO-2 and SUMO-3 under our conditions and that the resulting peptides are detected at least as well as those of SUMO-2/3 by LC/MS/MS. To determine what molar ratio of topo I to SUMO-1 would allow detection, we carried out spiking experiments using LC/MS/MS. SUMO-1 was detected in both trypsin and chymotrypsin digestions when 0.24 and 0.49 µg (respectively) was spiked into 4µg of purified CptTop1cc. These numbers correspond to molar ratios of 1:2 (SUMO-1:topo I) for trypsin, and 1:1 (SUMO-1:topo I) for chymotrypsin. Topo I and SUMO-2 were detected in all the samples. This result suggests that we should be able to detect SUMO-1 in our original experiments on purified CptTop1cc if there were as little as 0.5 SUMO-1 modification per topo I molecule (trypsin experiments) or one SUMO-1 modification per topo I (chymotrypsin experiments). Since slightly over half of the topo I in our preparations is sumoylated, we should detect SUMO-1 if the sumoylated fraction of CptTop1cc has as little as one SUMO-1 residue per sumoylated topo I molecule in CptTop1cc.
Western blotting
Western blotting was carried out on purified CptTop1cc and cell lysates using antibodies to ubiquitin, SUMO-2/3, SUMO-1 and topoisomerase I (Supporting Figure 5S). Western blotting of a whole cell extract detected a distinct band at ~90 kDa, the position of RanGAP1, an abundant protein known to be modified by SUMO-1. The SUMO-2/3 Western blot showed the pattern expected if the laddering of topo I from CptTop1cc is due to SUMO-2/3 modification. SUMO-1 Western blotting of identically isolated CptTop1cc showed no bands. Western blotting of a whole cell extract showed numerous bands of ubiquitin-positive proteins. The ubiquitin Western blotting of the purified CptTop1cc from cells treated with CPT at 37°C showed that the first topoisomerase I band at ~100 kDa and the topoisomerase I sumoylation bands were ubiquitin-positive. ATP depletion was done as described by Sorensen et al. (45) before addition of CPT and isolation of CptTop1cc. The CptTop1cc from ATP depleted cells showed no laddering, but the main topoisomerase I band remained ubiquitin-positive. CptTop1cc from heat shocked cells (42°C) also showed no laddering, but the main topoisomerase I band became more ubiquitin positive. Western blotting with anti-human topoisomerase I showed a single topoisomerase I band at ~100 kDa in whole cell extract. The topoisomerase I Western blot of purified CptTop1cc from cells treated with CPT at 37°C showed a complete topoisomerase I ladder. CptTop1cc from heat shocked cells and ATP depleted cells appeared similar, with loss of laddering due to sumoylation.
DISCUSSION
Our analysis of purified, endogenous CptTop1cc formed in CPT-treated human cells gave extensive sequence coverage for topo I and SUMO-2/3, and confirmed the presence of ubiquitin on CptTop1cc (18). In contrast to SUMO-1, SUMO-2 and SUMO-3 contain internal sumoylation consensus sites which allow them to form branching structures (33). Thus, a protein target could be modified by many SUMO-2/3 residues in a chain attached to a single sumoylation site. The only peptide unique to one of these two paralogs was the TENNDHINLK peptide unique to SUMO-2. Interestingly, it has been suggested that SUMO-3 (Swiss-Prot accession # P55854) forms polymeric chains more readily than SUMO-2 (Swiss-Prot accession # P61956) (33). However, we cannot rule out the possibility that the SUMO chains on CptTop1cc are mixed SUMO-2/3 chains. No peptides of topoisomerase II or SUMO-1 were detected by either MALDI-TOF MS or LC/MS/MS. Gel analyses of purified CptTop1cc and control samples from solvent treated cells show no evidence of topoisomerase II, which together with the absence of topo II peptides indicates that topoisomerase II complexes are below the levels detectable by gels or mass spectrometry. Heat shock prevents sumoylation of CptTop1cc (23), and no SUMO peptides were detected from CptTop1cc in our heat shocked samples. Proteomic studies of poly-SUMO-1 chains formed in vitro show that SUMO-1 chains digest well with trypsin (36), so poor digestibility of such chains seems unlikely to account for the absence of SUMO-1 peptides.
It has been suggested that SUMO-1 may cap polymeric chains of SUMO-2/3, acting as a chain terminator (32) and some evidence that this can occur has been obtained with an in vitro - in vivo SUMO conjugation experiment in mammalian cells extracts (46). Our data does not rule out such caps at levels below 0.5 SUMO-1 per topo I molecule (or 1 SUMO-1 residue per topo I molecule in the sumoylated fraction). Also, the N-terminal threonine of the TENNDHINLK peptide unique to SUMO-2 results from tryptic cleavage at K11, the lysine modified by isopeptide bond formation in SUMO-2/3 chains. Since isopeptide bond formation at this lysine would block cleavage at this site by trypsin, it seems likely that at least some SUMO-2 residues in the topoisomerase I DNA cleavage complex are neither branched nor capped. The high levels of SUMO chain formation on some fractions of topo I in CptTop1cc also suggest that chain formation is not being efficiently terminated by SUMO-1 capping. This makes sense in terms of the earlier observation that cells contain about four times more SUMO-2/3 than SUMO-1, and that most of the SUMO-1 is conjugated to RanGap1 (Ran GTPase-activating protein 1) in vivo (8). In in vitro experiments, it was shown that the SUMO ligase Ubc9 does not discriminate between SUMO paralogs, and in vivo experiments with a tagged substrate having a single sumoylation site demonstrated that SUMO-1 formed a mono-adduct, while SUMO-3 (Swiss-Prot accession # P55854) formed chains (33). It has also been suggested that SUMO-1 capping of SUMO-2/3 chains may inhibit their degradation (47).
A number of published reports have found SUMO-1 modifications of topo I. These studies have used either immunological or proteomic detection methods, and typically involve over-expression of proteins or expression of tagged proteins. Studies of in vivo CptTop1cc have been done with Western blotting of whole cell extracts, or with immunoprecipitates from these extracts. Single immunoprecipitations, even with antibodies directed against tagged protein targets, can be heavily contaminated with spurious proteins (48), and Western blotting, even when directed against tagged antigens, is only semi-quantitative. Since each Coomassie stained gel band contains > 0.1 µg of protein, the CptTop1cc sumoylation ladder bands might be positive in a SUMO-1 Western blot with SUMO-1 making up far less than 1% of the SUMO residues. In addition, both overexpression of SUMO paralogs or expression of SUMO forms with bulky tags can alter patterns of sumoylation (7,49,50), thus, the possible perturbation of the systems by overexpression of tagged SUMO paralogs remains a possibility. Our Western blots of purified CptTop1cc detected SUMO-2/3 and topoisomerase I but did not detect SUMO-1.
Global searches for sumoylation targets have been done, often using proteomic analysis and tagged SUMO paralogs that facilitate isolation of sumoylated proteins. To our knowledge, two of these studies have reported topo I to be a SUMO-1 target. One study used anti-HA-beads to isolate SUMO-conjugated proteins in cells transiently transfected with vectors expressing HA-tagged (hemagglutinin fusion tagged) SUMO-1 (51). Since single affinity tag isolations tend to have high levels of contaminating proteins (48), a tandem affinity purification, which greatly reduces contaminating proteins, was used in another study which found evidence of topo I as a target of both SUMO-1 and SUMO-3 (52). The latter study involved stressing of the cells with heat shock and treatment with MG-132 to increase levels of proteins conjugated to His-S-double tagged proteins expressing the SUMO-1 or SUMO-3 paralogs, and the authors acknowledge that spurious co-purifications are still probable. Neither of these studies employed camptothecin. In the absence of CPT, topo I from mammalian cells migrates as a single band at a position corresponding to 100 kDa and this is also true of topo I expressed in baculovirus or yeast (53). Heat shock blocks sumoylation of topo I in CPT treated cells, preventing the CPT-induced high molecular weight ladder of sumoylated topo I bands, but without affecting the density or electrophoretic migration of the 100 kDa topoisomerase I band (23,44). Thus, topo I is not thought to be sumoylated in the absence of CPT or other topo I poisons. Two other searches for SUMO-1 targets did not find topo I (38,54). A study of SUMO-3 (SwissProt # P55854) target proteins did not identify topo I as a target of SUMO-3 conjugation in the absence of CPT (55), and a SILAC (Stable Isotope Labeling with Amino acids in Cell culture) study of SUMO-1 and SUMO-3 (SwissProt P55854) targets did not identify topo I as a target of either (9). As with the other studies, the latter two did not employ camptothecin, consistent with the idea that topo I is not normally sumoylated.
Our Western blotting experiments on purified CptTop1cc indicated the presence of SUMO-2/3 in the topoisomerase I sumoylation bands, but did not detect SUMO-1. Although the ubiquitinated and sumoylated topo I released from the DNA of CPT treated cells may represent different populations of CptTop1cc, it is also possible that both modifications are on the same topo I molecules. SUMO-dependent ubiquitination is known to occur in some systems (reviewed in: 56), so it is possible that sumoylation of topo I in CptTop1cc drives subsequent ubiquitination. It is also possible that the SUMO-2/3 chains are themselves ubiquitinated (47), or even capped by ubiquitin. Ubiquitination of all of the CptTop1cc ladder bands was detected by Western blotting, including the most rapidly migrating band assumed not to be sumoylated. As discussed in the introduction, there is evidence that the sumoylation of topo I in CptTop1cc is involved in down-regulation of topo I and that this down-regulation is an important determinant of cancer cell sensitivity to camptothecins. We speculate that our gradient gel evidence for release of topo I proteolytic fragments from DNA of CPT treated cells may represent down-regulation of topoisomerase I by degradation of sumoylated and ubiquitinated CptTop1cc during CPT treatment of the cells. Heat shock and ATP depletion resulted in CptTop1cc without sumoylation ladder bands as detected by anti-SUMO-2/3 and anti-topoisomerase I Western blots. ATP depletion is expected to stop ATP-dependent conjugation of ubiquitin family proteins to their substrates, but would not be expected to inhibit ATP-independent SUMO hydrolases. Heat shock appeared to intensify ubiquitination of the un-sumoylated CptTop1cc.
Our study is the first biochemical analysis of highly purified, endogenous CptTop1cc. The cells used in our study were neither over-expressing SUMO paralogs, nor expressing tagged SUMO forms, the cells were not stressed or treated with proteasome inhibitors, and the CptTop1cc were purified away from other cellular proteins. Our evidence shows that CptTop1cc from camptothecin treated human breast cancer cells are heavily modified by SUMO-2 or SUMO-2/3.
If present, SUMO-1 residues must be lower than 0.5 per topo I molecule in CptTop1cc. At this low level, SUMO-1 capping seems unlikely to be significantly restricting the growth of branching SUMO-2/3 chains.
Supplementary Material
Gradient gel electrophoretic analysis of purified CptTop1cc, Western blotting of purified CptTop1cc for topoisomerase I, SUMO-2/3, SUMO-1, and ubiquitin, heat shock experiments, figures and tables of sequence ion assignments for peptides from recombinant SUMO paralogs and SUMO peptides identified in purified camptothecin-stabilized topoisomerase-DNA complexes. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENTS
We thank Dr. Arthur Haas for the generous gift of anti-ubiquitin antibody, Dr. Murugesan Rajaram for technical assistance, and the anonymous Biochemistry reviewers for their careful work and useful comments.
Abbreviations
- SUMO
Small Ubiquitin-like MOdifier
- E1
SUMO-activating enzyme
- E2
SUMO carrier protein
- topo I
topoisomerase I
- CPT
camptothecin
- CPTs
camptothecins
- CptTop1cc
topoisomerase I-DNA cleavage complex
- CsCl
cesium chloride
- DTT
dithiothreitol
- Nano-LC/MS/MS
Capillary-liquid chromatography-nanospray tandem mass spectrometry
- MALDI-TOF MS
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- ACTH
adrenocorticotropic hormone
- Glu-Fib
[Glu1]-Fibrinopeptide B
- LC
liquid chromatography
- LTQ
linear quadrupole ion trap
- MOWSE
MOlecular Weight SEarch
- MS/MS
tandem mass spectrometry
- SILAC
Stable Isotope Labeling with Amino acids in Cell culture
- MG-132
Z-Leu-Leu-Leu-aldehyde
Footnotes
This work was supported by NIH/NCI RO1-CA097107 to RMS and the Ohio State University Comprehensive Cancer Center.
REFERENCES
- 1.Jentsch S, Pyrowolakis G. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
- 2.Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 3.Johnson ES. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 4.Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106–4113. doi: 10.1242/jcs.038570. [DOI] [PubMed] [Google Scholar]
- 5.Dohmen RJ. SUMO protein modification. Biochim. Biophys. Acta. 2004;1695:113–131. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 9.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 11.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Zhu S, Guzzo CM, Ellis NA, Sung KS, Choi CY, Matunis MJ. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J Biol Chem. 2008;283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerscher O. SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer L, Ratain MJ. Clinical pharmacology of camptothecins. Ca. Chemother. Pharmacol. 1998;42:S31–S43. doi: 10.1007/s002800051077. [DOI] [PubMed] [Google Scholar]
- 15.Hsiang Y-H, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 16.Snapka RM. Catastrophic failure of DNA replication forks: structural and recombinational pathways. In: Snapka RM, editor. The SV40 replicon model for analysis of anticancer drugs. San Diego: Academic Press; 1996. pp. 39–64. [Google Scholar]
- 17.Snapka RM. Topoisomerase inhibitors can selectively interfere with different stages of simian virus 40 DNA replication. Mol. Cell. Biol. 1986;6:4221–4227. doi: 10.1128/mcb.6.12.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai SD, Liu LF, Vazquez-Abad D, D'Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J. Biol. Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- 19.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. Proc. Natl. Acad. Sci. USA. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, Liu LF. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 2001;61:5926–5932. [PubMed] [Google Scholar]
- 21.Fu Q, Kim SW, Chen HX, Grill S, Cheng YC. Degradation of topoisomerase I induced by topoisomerase I inhibitors is dependent on inhibitor structure but independent of cell death. Mol. Pharmacol. 1999;55:677–683. [PubMed] [Google Scholar]
- 22.Horie K, Tomida A, Sugimoto Y, Yasugi T, Yoshikawa H, Taketani Y, Tsuruo T. SUMO-1 conjugation to intact DNA topoisomerase I amplifies cleavable complex formation induced by camptothecin. Oncogene. 2002;21:7913–7922. doi: 10.1038/sj.onc.1205917. [DOI] [PubMed] [Google Scholar]
- 23.Mo YY, Yu Y, Shen Z, Beck WT. Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J. Biol. Chem. 2002;277:2958–2964. doi: 10.1074/jbc.M108263200. [DOI] [PubMed] [Google Scholar]
- 24.Rallabhandi P, Hashimoto K, Mo YY, Beck WT, Moitra PK, D'Arpa P. Sumoylation of topoisomerase I is involved in its partitioning between nucleoli and nucleoplasm and its clearing from nucleoli in response to camptothecin. J. Biol. Chem. 2002;277:40020–40026. doi: 10.1074/jbc.M200388200. [DOI] [PubMed] [Google Scholar]
- 25.Walters KJ, Goh AM, Wang Q, Wagner G, Howley PM. Ubiquitin family proteins and their relationship to the proteasome: a structural perspective. Biochim. Biophys. Acta. 2004;1695:73–87. doi: 10.1016/j.bbamcr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Mo YY, Wang P, Beck WT. Functional expression of human DNA topoisomerase I and its subcellular localization in HeLa cells. Exp. Cell Res. 2000;256:480–490. doi: 10.1006/excr.2000.4864. [DOI] [PubMed] [Google Scholar]
- 27.Christensen MO, Barthelmes HU, Feineis S, Knudsen BR, Andersen AH, Boege F, Mielke C. Changes in mobility account for camptothecin-induced subnuclear relocation of topoisomerase I. J. Biol. Chem. 2002;277:15661–15665. doi: 10.1074/jbc.C200066200. [DOI] [PubMed] [Google Scholar]
- 28.Christensen MO, Barthelmes HU, Boege F, Mielke C. The N-terminal domain anchors human topoisomerase I at fibrillar centers of nucleoli and nucleolar organizer regions of mitotic chromosomes. J. Biol. Chem. 2002;277:35932–35938. doi: 10.1074/jbc.M204738200. [DOI] [PubMed] [Google Scholar]
- 29.Jacquiau HR, van Waardenburg RC, Reid RJ, Woo MH, Guo H, Johnson ES, Bjornsti MA. Defects in SUMO (small ubiquitin-related modifier) conjugation and deconjugation alter cell sensitivity to DNA topoisomerase I-induced DNA damage. J. Biol. Chem. 2005;280:23566–23575. doi: 10.1074/jbc.M500947200. [DOI] [PubMed] [Google Scholar]
- 30.Beidler DR, Cheng Y-C. Camptothecin induction of a time- and concentration-dependent decrease of topoisomerase I and its implication in camptothecin activity. Mol. Pharmacol. 1995;47:907–914. [PubMed] [Google Scholar]
- 31.Danks MK, Garrett KE, Marion RC, Whipple DO. Subcellular redistribution of DNA topoisomerase I in anaplastic astrocytoma cells treated with topotecan. Cancer Res. 1996;56:1664–1673. [PubMed] [Google Scholar]
- 32.Yang M, Hsu CT, Ting CY, Liu LF, Hwang J. Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. J. Biol. Chem. 2006;281:8264–8274. doi: 10.1074/jbc.M510364200. [DOI] [PubMed] [Google Scholar]
- 33.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 34.Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 2002;277:47938–47945. doi: 10.1074/jbc.M207442200. [DOI] [PubMed] [Google Scholar]
- 35.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 36.Pedrioli PG, Raught B, Zhang XD, Rogers R, Aitchison J, Matunis M, Aebersold R. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat. Methods. 2006;3:533–539. doi: 10.1038/nmeth891. [DOI] [PubMed] [Google Scholar]
- 37.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 38.Gocke CB, Yu H, Kang J. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J. Biol. Chem. 2005;280:5004–5012. doi: 10.1074/jbc.M411718200. [DOI] [PubMed] [Google Scholar]
- 39.Hammer E, Heilbronn R, Weger S. The E3 ligase Topors induces the accumulation of polysumoylated forms of DNA topoisomerase I in vitro and in vivo. FEBS Lett. 2007;581:5418–5424. doi: 10.1016/j.febslet.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 40.Pappin DJ, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 41.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Crystal structure of SUMO-3-modified thymine-DNA glycosylase. J. Mol. Biol. 2006;359:137–147. doi: 10.1016/j.jmb.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 42.Shen LN, Dong C, Liu H, Naismith JH, Hay RT. The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing. Biochem. J. 2006;397:279–288. doi: 10.1042/BJ20052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haas AL, Bright PM. The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J Biol Chem. 1985;260:12464–12473. [PubMed] [Google Scholar]
- 44.Mao Y, Desai SD, Liu LF. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem. 2000;275:26066–26073. doi: 10.1074/jbc.M001831200. [DOI] [PubMed] [Google Scholar]
- 45.Sorensen M, Sehested M, Jensen PB. Effect of cellular ATP depletion on topoisomerase II poisons. Abrogation of cleavable-complex formation by etoposide but not by amsacrine. Mol. Pharmacol. 1999;55:424–431. [PubMed] [Google Scholar]
- 46.Matic I, van HM, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell Proteomics. 2008;7:132–144. doi: 10.1074/mcp.M700173-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petrie K, Zelent A. Marked for death. Nat. Cell Biol. 2008;10:507–509. doi: 10.1038/ncb0508-507. [DOI] [PubMed] [Google Scholar]
- 48.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 49.Kamitani T, Kito K, Nguyen HP, Fukuda-Kamitani T, Yeh ET. Characterization of a second member of the sentrin family of ubiquitin-like proteins. J. Biol. Chem. 1998;273:11349–11353. doi: 10.1074/jbc.273.18.11349. [DOI] [PubMed] [Google Scholar]
- 50.Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., III Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Y, Kwon SW, Anselmo A, Kaur K, White MA. Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J. Biol. Chem. 2004;279:20999–21002. doi: 10.1074/jbc.M401541200. [DOI] [PubMed] [Google Scholar]
- 52.Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell Proteomics. 2005;4:56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soe K, Dianov G, Nasheuer HP, Bohr VA, Grosse F, Stevnsner T. A human topoisomerase I cleavage complex is recognized by an additional human topoisomerase I molecule in vitro. Nucleic Acids Res. 2001;29:3195–3203. doi: 10.1093/nar/29.15.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, Stadtman ER, Yang DC, Chock PB. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl. Acad Sci U. S. A. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 56.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gradient gel electrophoretic analysis of purified CptTop1cc, Western blotting of purified CptTop1cc for topoisomerase I, SUMO-2/3, SUMO-1, and ubiquitin, heat shock experiments, figures and tables of sequence ion assignments for peptides from recombinant SUMO paralogs and SUMO peptides identified in purified camptothecin-stabilized topoisomerase-DNA complexes. This material is available free of charge via the Internet at http://pubs.acs.org.