Abstract
CST6 is a breast tumor suppressor gene that is expressed in normal breast epithelium, but is epigenetically silenced as a consequence of promoter hypermethylation in metastatic breast cancer cell lines. In the current study, we investigated the expression and methylation status of CST6 in primary breast tumors and lymph node metastases. 25/45 (56%) primary tumors and 17/20 (85%) lymph node metastases expressed significantly lower levels of cystatin M compared to normal breast tissue. Bisulfite sequencing demonstrated CST6 promoter hypermethylation in 11/23 (48%) neoplastic lesions analyzed, including 3/11 (27%) primary tumors and 8/12 (67%) lymph node metastases. In most cases (12/23, 52%), the expression of cystatin M directly reflected CST6 promoter methylation status. In remaining lesions (8/23, 35%) loss of cystatin M was not associated with CST6 promoter hypermethylation, indicating that other mechanisms can account for loss of CST6 expression. These results show that methylation-dependent silencing of CST6 occurs in a subset of primary breast cancers, but more frequently in metastatic lesions, possibly reflecting progression-related genomic events. To examine this possibility, primary breast tumors and matched lymph node metastases were analyzed. In 2/3 (67%) patients, primary tumors were positive for cystatin M and negative for CST6 promoter methylation, and matched metastatic lesions lacked cystatin M expression and CST6 was hypermethylated. This observation suggests that progression-related epigenetic alterations in CST6 gene expression can accompany metastatic spread from a primary tumor site. Overall, the results of the current investigation suggest that methylation-dependent epigenetic silencing of CST6 represents an important mechanism for loss of CST6 during breast tumorigenesis and/or progression to metastasis.
Keywords: breast cancer, cystatin M, methylation, metastasis
Introduction
Breast cancer is a heterogeneous disease that results from the accumulation of a complex series of genetic and epigenetic events driving divergent pathways that ultimately convey varying phenotypic properties to individual neoplastic lesions. Most invasive breast cancers (approximately 90%) are of the ductal or lobular histopathological type (Symmans, 2005). Numerous molecular markers have been examined for their predictive value in breast cancer prognostication, but histopathologic grade emerges as the most important indicator of long-term patient outcome (Simpson et al., 2005; Symmans, 2005). However, histopathologic grade generally correlates with the expression of genes associated with increased cell proliferation (Ki-67, p53), growth (HER-2), and invasiveness (matrix metalloproteinases) (Mirza et al., 2002; Reed et al., 2000). In contrast, low-grade breast tumors express genes associated with low cellular proliferation (p27) and differentiation (ER and PR) (Lau et al., 2001; Symmans, 2005). It is now accepted that there are two major pathways of multi-step breast cancer progression, (i) well-differentiated ductal carcinoma in situ (DCIS) progressing to grade I invasive ductal carcinoma (IDC), and (ii) poorly differentiated DCIS progressing to grade III IDC (Simpson et al., 2005). Changes in the molecular pattern of DCIS lesions may lead to the ability to collapse the myoepithelium, escape the duct structure, and invade the surrounding stroma (Ai et al., 2006; Symmans, 2005). These IDC lesions will proliferate and grow, destroying surrounding stroma, and breast architecture. Continued disease progression can lead to tumor dissemination via lymphatic or hematogenous routes giving rise to metastatic lesions in distant organs (Muller et al., 2005). At present, the molecular mechanisms that control tumor progression, stromal invasion, and distant metastasis are poorly understood. Nevertheless, the role of specific genes that contribute to breast cancer tumor invasion and metastasis are beginning to be investigated and characterized.
Cystatin M (CST6) functions as an endogenous inhibitor of lysosomal cysteine proteases (Ni et al., 1997; Sotiropoulou et al., 1997), which are known to contribute to tumor cell invasion by degrading extracellular matrix components (Buck et al., 1992; Maciewicz et al., 1990; Mai et al., 2002). Cathepsin B and cathepsin L are two important cysteine proteases that have been implicated in tumor cell invasion and metastasis (Kane and Gottesman, 1990; Roshy et al., 2003; Slone et al., 1981). Cystatin M is involved in regulating the activity of cathepsin B and cathepsin L, and imbalances between these proteases and cystatin M may lead to the metastatic phenotype in tumor cells (Frosch et al., 1999; Kos et al., 2000; Lah and Kos, 1998). Cystatin M expression has been reported to be diminished or lost in various forms of cancer including, (i) basal and squamous cell carcinomas of the skin (Zeeuwen, 2004), (ii) squamous cell carcinomas of the head/neck and lung (Zeeuwen et al., 2002), (iii) non-small cell lung cancer (Zhong et al., 2006), (iv) metastatic oral cancer cell lines (Vigneswaran et al., 2006), (v) malignant glioma (Kim et al., 2006), and (vi) breast cancer (Ai et al., 2006; Rivenbark et al., 2006a; Schagdarsurengin et al., 2006; Shridhar et al., 2004; Song et al., 2006; Sotiropoulou et al., 1997; Zhang et al., 2004). Cystatin M contains a large CpG island that flanks the transcription start site and spans the proximal promoter and exon 1 regions. CpG islands represent well-characterized sites for DNA methylation, impairing transcription activation related to regional or specific methylation events (Deng et al., 2004; Douglas et al., 2004; Prendergast and Ziff, 1991). In a previous microarray-based analysis of differential gene expression in MCF-7 breast cancer cells, we identified CST6 as a methylation-sensitive gene, and demonstrated an inverse relationship between CST6 mRNA expression and methylation of the proximal promoter region of its CpG island (Rivenbark et al., 2006b). In a subsequent study, methylation analysis of CST6 in breast cancer cell lines revealed an inverse correlation between CpG island hypermethylation and CST6 gene expression, and the extent of regional methylation in the proximal promoter was strongly correlated with the lack of CST6 expression (Rivenbark et al., 2006a). Consistent with these findings, several other studies have shown that CST6 is epigenetically regulated by DNA methylation-dependent silencing in breast cancer cell lines and primary invasive ductal carcinomas (Ai et al., 2006; Schagdarsurengin et al., 2006).
The aim of this study was to investigate cystatin M protein expression and CST6 gene methylation status in primary breast cancers and lymph node metastasis in order to determine if aberrant DNA methylation represents an important mechanism for the loss of CST6 during neoplastic transformation of breast epithelium and/or breast cancer progression. Immunochemical analysis revealed 25/45 (56%) primary breast tumors and 17/20 (85%) lymph node metastases express significantly lower levels of cystatin M protein compared to normal breast tissue. Bisulfite sequencing demonstrated CST6 promoter hypermethylation in 3/11 (27%) primary breast tumors and 8/12 (67%) lymph node metastases analyzed. In most cases (12/23, 52%), the extent of CST6 promoter methylation was associated with the expression of cystatin M protein. In other neoplastic lesions, (8/23, 35%) loss of cystatin M expression in the absence of CST6 promoter methylation indicates that other mechanisms can account for silencing of CST6. These results show that methylation-dependent epigenetic silencing of CST6 occurs in a subset of primary breast tumors, but more frequently in metastatic lesions, possibly reflecting the role of cystatin M in breast cancer progression/invasion. To examine the possibility that CST6 is silenced by methylation during tumor progression, matched primary breast tumors and lymph node metastases were analyzed. In our limited analysis, the majority (2/3, 67%) of cystatin M-positive primary tumors (lacking CST6 promoter methylation) give rise to cystatin M-negative lymph node metastases with hypermethylation of the CST6 promoter. Combined, the observations of the current study suggests strongly that CST6 promoter hypermethylation and loss of cystatin M expression affects a subset of human breast cancers and that methylation-dependent epigenetic silencing of CST6 can occur during breast tumorigenesis (early) or progression (late), contributing to tumor metastasis.
Materials and Methods
Human Breast and Lymph Node Tissues
This study included 87 paraffin-embedded human tissues corresponding to primary breast tumors (n=54), lymph nodes metastases (n=22), and normal breast tissues (n=11). Twenty-one archival human tissues (primary breast tumors, lymph node metastases, and normal breast) were obtained from the University of North Carolina Lineberger Comprehensive Cancer Center and 6 archival primary breast tumors were acquired from the Louisiana State University Health Sciences Center. A breast tumor microarray (Imgenex Corporation, Sorrento Valley, CA) consisting of 60 tissue cores was also utilized. In total, this study included 46 primary breast specimens diagnosed invasive ductal carcinoma (IDC), 2 breast ductal carcinoma in situ specimens, 1 solid papillary carcinoma, 1 medullary carcinoma, 1 signet ring cell carcinoma, 3 infiltrating lobular carcinomas, 22 lymph node metastases from IDC (n=20), atypical medullary carcinoma (n=1), and infiltrating lobular carcinoma (n=1), and 11 normal breast tissue samples. Five archival primary breast tumors were matched paired with lymph node metastases. Handling of tissue specimens and protection of patient privacy followed strict policies of the institutional review board of the University of North Carolina School of Medicine.
Immunohistochemistry
Formalin-fixed, paraffin-embedded human breast tissues and lymph nodes were sectioned (5 μm thick) and mounted on glass microscope slides. Immunohistochemical staining was performed according to standard methods. Briefly, tissue sections were incubated on a slide warmer at 60°C for 15 minutes, deparaffinized in xylene, incubated with 3% H2O2 in methanol to block endogenous peroxidase activity, and rehydrated through a series of ethanol washes. Antigen retrieval was accomplished by steaming in 1X citrate buffer (Dako Inc., Carpinteria, CA) for 30 minutes. After incubation with serum-free protein block (Dako Inc.) for 10 minutes, tissues were incubated for 2 hours at room temperature with polyclonal rabbit anti-cystatin M antibodies diluted 1:1000 (Zhang et al., 2004). Subsequently, tissues were washed and layered with a two-step secondary set-up including a anti-rabbit biotintylated link and streptavidin-conjugated HRP solution (Dako Inc.) for 10 minutes each, incubated with HRP substrate containing 3,3′ diaminobenzidine (Dako Inc.) for a total of 5 minutes, followed by counterstaining with hematoxylin. Control immunostaining reactions were performed at room temperature with mouse monoclonal anti-cytokeratin 18 (Santa Cruz Biotechnology Inc., Santa Cruz, CA) antibodies diluted 1:1000. Negative control staining followed the same procedure except sections were incubated with either rabbit preimmune serum or 1x wash buffer instead of anti-cystatin M antibody. Normal breast tissue was used as a positive control for the anti-cystatin M antibody.
Isolation of Genomic DNA from Human Primary Breast Tumors, Lymph Nodes, and Normal Breast Tissue
Paraffin-embedded tissue specimens were scraped or microdissected from slides using a clean razor blade, deparaffinized, and genomic DNA was isolated using a QIAamp DNA Micro kit (Qiagen, Inc., Valencia, VA) according to manufacturer’s instructions. Briefly, tissue samples were incubated overnight at 56°C with proteinase K. Subsequently, carrier RNA (1 μg/μl) was added and DNA samples were applied to columns, washed, and eluted with 35 μl of distilled water.
Sodium Bisulfite Modification of Genomic DNA, Cloning, and Sequencing
Genomic DNA was modified with sodium bisulfite using the EZ DNA Methylation-Gold Kit (ZYMO Research Co., Orange, CA) according to the manufacturer’s protocol. Briefly, approximately 1.5 μg denatured genomic DNA was treated with conversion reagent, incubated at 98°C for 10 minutes, and 53°C for 30 minutes, followed by a step-cycle program consisting of 8 cycles of 53°C for 6 minutes and 37°C for 30 minutes. Subsequently, samples were applied to columns, washed, desulfonated, washed and then eluted with 20 μl of elution buffer. In general, 2 μl of modified DNA was used in subsequent PCR reactions. Sodium bisulfite-converted DNA was amplified using primers that targeted a region of the proximal promoter and exon 1 of CST6 encompassing nucleotides −118 to +242, which contains 46 CpG dinucleotides (5′-GGTTTTTTGGGTTTTTTGAATTT-3′ and 5′-CTACCCATATTATAACTAACC-3′). PCR amplification was accomplished using a step-cycle program consisting of 40 cycles of 94°C for denaturing (1 minute), 52°C for annealing (1.5 minute), and 72°C for extension (2 minutes). PCR products were fractionated on 2% agarose gels containing 40 mM Tris-acetate/1.0 mM EDTA (pH 8.0) and visualized by ethidium bromide staining. A portion of each PCR product (1 to 5 μl) was cloned into pGEM-T Easy Vector (Promega, Madison, WI) using standard methods. Five to 12 colonies corresponding to each tissue sample were selected and expanded in liquid culture. Plasmid DNA was purified using the Wizard Plus SV Miniprep DNA purification kit (Promega), prior to digestion with NcoI and NdeI (New England Biolabs, Beverly, MA) to confirm the presence and size of the cloned insert. Validated clones were sequenced using the universal M13R3 primer with an Applied Biosystems automated sequencer at the UNC Genome Analysis Facility (Chapel Hill, NC). The bisulfite conversion efficiency was calculated for each sequenced clone based upon the ratio of converted Cs (non-CpG) to total number of Cs (non-CpG) in a given gene segment. Only clones determined to have a conversion efficiency of >95% were included in the present study. The results of methylation analyses were expressed as total methylation index (TMI). This measure of methylation can be applied to single CpG dinucleotides, select groups of CpG dinucleotides, or to continuous groups of CpG dinucleotides in a given gene segment. TMI was calculated for each tumor and clone by dividing the number of methylated CpGs observed by the total CpGs analyzed and expressed as percent methylation (Rivenbark et al., 2006a). For instance, in an analysis containing 46 CpG dinucleotides and 12 clones sequenced, TMI would be calculated based upon 552 possible CpG methylation events (12 x 46). Tumors with a CST6 promoter TMI >11% were considered hypermethylated.
Statistical Analysis
Values included in the text, tables, and figures represent averages ± S.E.M. that were calculated using the statistical function of KaleidaGraph Version 3.5 (Synergy Software, Essex Junction, VT). An unpaired t-test was used to examine the association between CST6 methylation status and protein expression levels among subsets of primary breast tumors, lymph node metastases, and normal breast.
Results
Immunohistochemical Analysis of Cystatin M in Primary Breast Tumors and Lymph Node Metastases
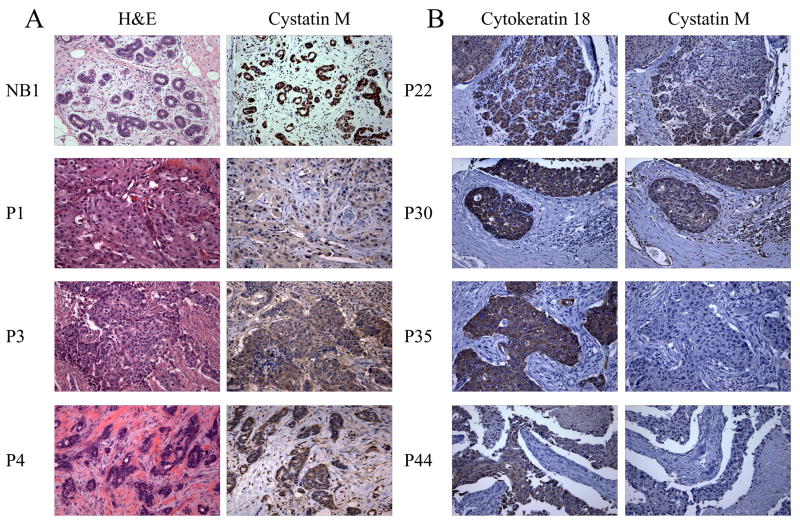
The expression of cystatin M was examined at the protein level using paraffin-embedded tissues and immunohistochemical (IHC) staining. Table 1 shows primary breast tumor designations along with tumor type, pTNM, pathological stage, and cystatin M protein expression status. A breast tissue microarray (TMA) containing 60 tissue cores was immunostained for cystatin M and cytokeratin 18 (CK18). Seventeen tissue cores from the TMA could not be scored due to an absence of CK18 immunostaining. The remaining 43/60 (72%) cores (including 31 primary tumors) showed strong staining for CK18. Therefore, these 43 tissue samples from the TMA were analyzed for cystatin M expression. Immunodetection of cystatin M in select normal human breast tissues (total n=5) and primary breast tumors (total n=45) are shown in Fig. 1 and summarized in Table 1. Epithelial and myoepithelial cells of 5/5 (100%) normal breast tissues showed strong immunostaining for cystatin M (Fig. 1A, NB1). Likewise, 20/45 (44%) primary tumors (Fig. 1, P3, P4, P22, and P30) were positive for cystatin M expression. In contrast, 25/45 (56%) primary breast tumors were found to be negative for cystatin M (Fig. 1, P1, P35 and P44), including 21/38 (55%) IDC, 1/2 (50%) ductal carcinoma in situ (DCIS), 1/1 (100%) solid papillary carcinoma, 1/3 (33%) infiltrating lobular carcinoma, and 1/1 (100%) signet ring cell carcinoma.
Table 1.
Characteristics of human primary breast tumors and normal breast tissues.
| Tissue Designation1 | Tumor Type2 | pTNM | Stage | Cystatin M Expression |
|---|---|---|---|---|
| P1 | IDC | T2N1Mx | IIB | No |
| P2 | IDC | T1cN3aMx | IIIC | Yes |
| P3 | IDC | T2N1Mx | IIB | Yes |
| P4 | IDC | T2N1Mx | IIB | Yes |
| P5 | IDC | T1cN2aMx | IIIA | No |
| P6 | IDC | T1NxMx | UNK | No |
| P7 | IDC | TxNxMx3 | IIA | Yes |
| P8 | IDC | T2N1Mx | IIB | No |
| P9 | IDC | T1bN0Mx | I | No |
| P10 | IDC | T1cN1Mx | IIA | No |
| P11 | IDC | T1N1Mx | IIA | No |
| P12 | IDC | T2N1Mx | IIB | No |
| P13 | IDC | T4N3M1 | IV | Yes |
| P14 | IDC | T1N0Mx | I | Yes |
| P15 | solid papillary carcinoma | T3N1aM0 | IIIA | No |
| P16 | IDC | T4bN2aM0 | IIIB | No |
| P17 | IDC | T3N0M0 | IIB | No |
| P18 | IDC | T3N1aM0 | IIIA | No |
| P19 | IDC | T3N3aN0 | IIIC | No |
| P20 | infiltrating lobular carcinoma | T2N1aM0 | IIB | Yes |
| P21 | IDC | T3N1aM0 | IIIA | Yes |
| P22 | IDC | T2N1aM0 | IIB | Yes |
| P23 | infiltrating lobular carcinoma | T3N3aM0 | IIIC | Yes |
| P24 | infiltrating lobular carcinoma | T3N3aM0 | IIIC | No |
| P25 | IDC | T2N1aM0 | IIB | No |
| P26 | IDC | T2N0M0 | IIA | No |
| P27 | DCIS | TisN0M0 | 0 | No |
| P28 | IDC | T2N2aM0 | IIIA | Yes |
| P29 | IDC | T2N3bM0 | IIIC | Yes |
| P30 | DCIS | TisN0M0 | 0 | Yes |
| P31 | IDC | T2N2M0 | IIIA | Yes |
| P32 | IDC | T2N0M0 | IIA | No |
| P33 | IDC | T2N0M0 | IIA | Yes |
| P34 | signet ring cell carcinoma | T3N0M0 | IIB | No |
| P35 | IDC | T3N2aM0 | IIIA | No |
| P36 | IDC | T3N2aM0 | IIIA | No |
| P37 | IDC | T2N0M0 | IIA | Yes |
| P38 | IDC | T2N0M0 | IIA | Yes |
| P39 | IDC | T2N2aM0 | IIIA | Yes |
| P40 | IDC | T2N0M0 | IIB | Yes |
| P41 | IDC | T3N1aM0 | IIIA | No |
| P42 | IDC | T2N1aM0 | IIB | Yes |
| P43 | IDC | T2N0M0 | IIA | No |
| P44 | IDC | T3N2aM0 | IIIA | No |
| P45 | IDC | T3N1aM0 | IIIA | No |
|
| ||||
| NB1 | Normal | NA | NA | Yes |
| NB2 | Normal | NA | NA | Yes |
| NB3 | Normal | NA | NA | Yes |
| NB4 | Normal | NA | NA | Yes |
| NB5 | Normal | NA | NA | Yes |
Primary breast tumors are indicated as Px and normal breast tissues are designated NBx.
IDC refers to invasive ductal carcinoma and DCIS refers in ductal carcinoma in situ.
TxNxMx refers to a pTNM that is unknown.
Fig. 1.
Immunohistochemical analysis of cystatin M expression in human primary breast tumors. (A) Panels show H&E and cystatin M immunostaining in the same tumors. Normal breast (NB1) and primary tumors P3 and P4 show positive staining for cystatin M. Tumor P1 shows reduced cystatin M staining compared to NB1. (B) Panels show cytokeratin 18 (CK18) and cystatin M immunostaining in the same tumors. All tumors show strong staining for CK18. Tumors P22 and P30 exhibit positive cystatin M immunostaining. Tumors P35 and P44 show reduced cystatin M staining. (Original objective lens magnification 10x).
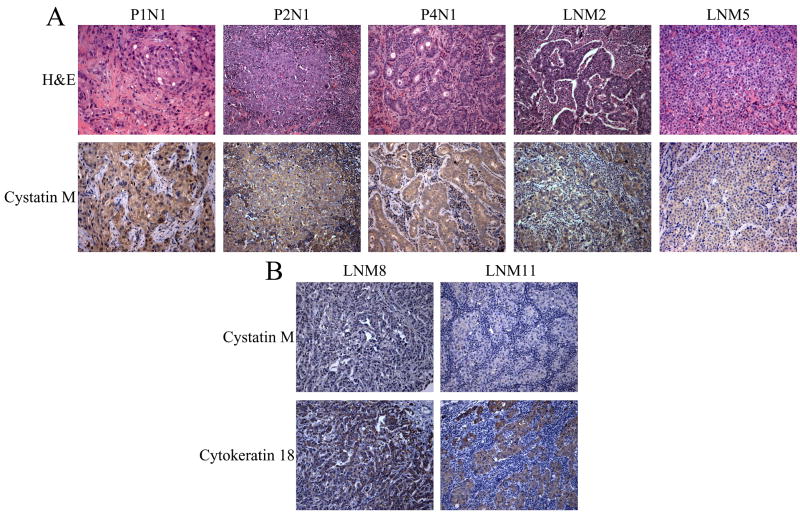
To examine cystatin M protein expression in metastatic lesions, 20 lymph nodes were obtained for immunostaining (12 of the lymph nodes were derived from 5 primary breast tumors). Table 2 contains information related to the lymph nodes analyzed, including tumor designations, along with tumor type, pTNM, pathological stage from matched primary tumors, and cystatin M protein expression status. Fig. 2 shows representative cystatin M IHC staining in these metastatic lesions. The majority (17/20, 85%) of metastatic lesions in lymph nodes were negative for cystatin M expression (Fig. 2, P2N1, P4N1, LNM2, LNM5, LNM8, and LNM11). In contrast, 3/20 (15%) metastatic lesions were positive for cystatin M expression (Fig. 2, P1N1). Overall, these numbers reflect the presence of cystatin M-negative lymph node metastases in 13/16 (81%) patients. These observations are consistent with the suggestion that loss of cystatin M expression is a common feature of metastatic breast tumors. However, the presence of cystatin M-positive breast tumor in some lymph node specimens suggests that loss of cystatin M expression is not absolutely required for tumor invasion and metastasis.
Table 2.
Characteristics of human lymph node metastases.
| Tissue Designation1 | Tumor Type | pTNM2 | Stage2 | Cystatin M Expression |
|---|---|---|---|---|
| P1N1 | lymph node metastasis | T2N1Mx | IIB | Yes |
| P2N1-N33 | lymph node metastasis | T1cN3aMx | IIIC | No |
| P3N1 | lymph node metastasis | T2N1Mx | IIB | Yes |
| P4N1 | lymph node metastasis | T2N1Mx | IIB | No |
| P5N1 | lymph node metastasis | T1cN2aMx | IIA | No |
| LNM1-44 | lymph node metastasis | TxNxMx | UNK | No |
| LNM5 | lymph node metastasis | TxNxMx | UNK | No |
| LNM6 | lymph node metastasis | T3N3aM0 | IIIC | Yes |
| LNM7 | lymph node metastasis | T3N3aM0 | IIIC | No |
| LNM8 | lymph node metastasis | T3N3aM0 | IIIC | No |
| LNM9 | lymph node metastasis | T3N3M0 | IIIC | No |
| LNM10 | lymph node metastasis | T3N2aM0 | IIIA | No |
| LNM11 | lymph node metastasis | T1cN1aM0 | IIA | No |
| LMN12 | lymph node metastasis | T2N3aM0 | IIIC | No |
| LMN13 | lymph node metastasis | T1bN3aM0 | IIIC | No |
Metastaic lesions corresponding to a matched primary breast tumor are indicated as PxNx and lesions that have no matched primary are designated LNMx.
pTNM and pathological stage designations for lymph node metastases are derived from primary breast tumor designations. TxMxNx refers to a pTNM that is unknown.
P2N1-N3 designates 3 independent lymph nodes corresponding to one primary tumor.
LNM1-4 designates 4 independent lymph nodes corresponding to one primary tumor.
Fig. 2.
Immunohistochemical analysis of cystatin M expression in lymph node metastases. (A) Panels show H&E and cystatin M immunostaining in the same lymph nodes. Lymph node P1N1 shows positive staining for cystatin M. Lymph nodes P2N1, P4N1, LNM2, and LNM5 show reduced cystatin M immunostaining. (B) Panels show cytokeratin 18 (CK18) and cystatin M immunostaining in the same lymph nodes. All metastatic lesions show strong staining for CK18 and exhibit reduced cystatin M immunostaining. (Original objective lens magnification 10x).
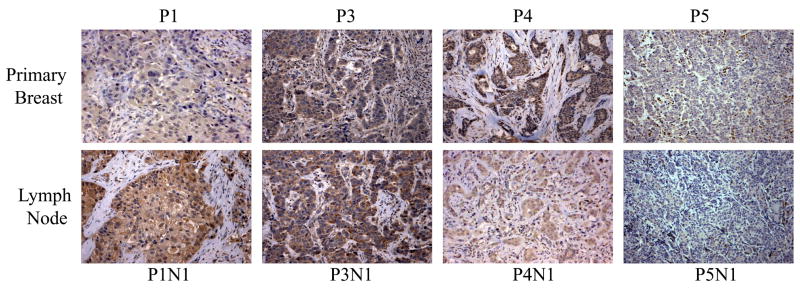
To examine the possibility that loss of cystatin M reflects a tumor progression-related event, 5 primary breast tumors and matched lymph node metastases were immunostained. Fig. 3 shows representative examples of these primary tumor/lymph node pairs. 1/5 (20%) matched pairs were negative for cystatin M expression in both the primary breast tumor and lymph node metastasis, indicating an early loss of cystatin M protein during breast tumorigenesis, with persistence in the metastatic lesion (Fig. 3, P5 and P5N1). Additionally, 1/5 (20%) matched pairs showed positive cystatin M staining in both the primary breast and lymph node tissues, suggesting that tumor metastasis in this patient was mediated through a cystatin M-independent pathway (Fig. 3, P3 and P3N1). The remaining 3 patients had cystatin M-positive primary tumors, but 2/3 (67%) of the matched lymph node metastases lacked cystatin M expression (Fig. 3, P4 and P4N1). This observation is consistent with a progression related loss of cystatin M expression during the evolution of the metastatic clone. The remaining primary tumor/lymph node pair lacks cystatin M expression in the primary tumor, while the lymph node metastasis exhibits stronger staining for the cystatin M protein (Fig. 3, P1 and P1N1). This result is unclear, but may reflect heterogeneity of cystatin M expression in this tumor. Thus, a cystatin M-positive tumor cell population may have given rise to this metastatic lesion through a cystatin M-independent pathway.
Fig. 3.
Immunohistochemical analysis of cystatin M expression in matched primary breast tumors and lymph node metastases. Representative examples of matched pairs of primary breast tumors (top panel) and lymph node metastasis (bottom panel) are shown. Primary breast tumor P1 shows reduced cystatin M immunostaining compared to its matched lymph node P1N1. Primary breast tumor P3 and lymph node metastasis P3N1 both show positive staining for cystatin M. Primary breast tumor P4 shows positive staining for cystatin M compared to its matched lymph node metastasis P4N1. Primary breast tumor P5 and lymph node metastasis P5N1 both show negative staining for cystatin M. (Original objective lens magnification 10x).
Methylation-Dependent Silencing of CST6 in Primary Breast Tumors and Lymph Node Metastases
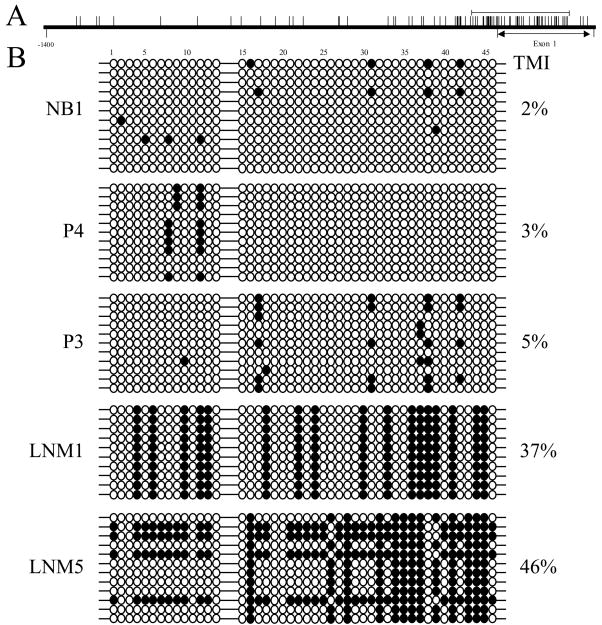
To explore the possibility that loss of cystatin M expression is related to epigenetic silencing of CST6, we analyzed primary breast tumors and lymph node metastases for CST6 promoter methylation. A segment of the proximal promoter and exon 1 (−118 to +242, Fig. 4A) containing 46 CpG dinucleotides was analyzed in normal breast tissue, 11 primary breast tumors (5 express cystatin M and 6 lack cystatin M), and 12 lymph node metastases (2 express cystatin M and 10 lack cystatin M). Multiple clones (n=5–12) corresponding to the CST6 promoter and exon 1 from each primary tumor or lymph node metastasis were analyzed by sodium bisulfite sequencing and individual CpGs were scored for methylation status. Representative examples are shown in Fig. 4B. In normal breast tissue, 33/46 (72%) CpGs were consistently unmethylated, 13/46 (28%) CpGs were methylated at an intermediate level, and 0/46 (0%) were 100% methylated, producing a TMI of 2% (Fig. 4B, NB1). The majority (3/5, 60%) of primary tumors that stain positive for cystatin M lack appreciable levels of methylation, with TMI values of 0 to 5% (Fig. 4B, P4 and P3). Primary breast tumors P2 and P9, exhibit methylation and express cystatin M. In P2, 23/46 (50%) CpGs were unmethylated, 22/46 (48%) CpGs were methylated at an intermediate level, and 1/46 (2%) was 100% methylated, producing a TMI of 38%. In P9, the majority (33/46, 72%) of CpGs were unmethylated, although 13/46 (28%) were 100% methylated resulting in a TMI of 28%. The continued expression of cystatin M in P2 and P9, despite CST6 promoter hypermethylation, suggests that there may be mechanisms to transcriptionally bypass promoter methylation. Most, (5/6, 83%) primary breast tumors that are negative for cystatin M expression exhibit very low levels of CST6 methylation (TMI = 0 to 3%). This finding suggests that there may be other epigenetic or mutational mechanisms responsible for the silencing of cystatin M in these primary breast tumors. In contrast, one tumor (P1) was negative for cystatin M protein expression and 45/46 (98%) CpGs were methylated at an intermediate level, resulting in a TMI of 28%. Overall, a subset of primary tumors (3/11, 27%) exhibits CST6 promoter hypermethylation, and in one case this methylation was associated with loss of cystatin M expression.
Fig. 4.
Methylation analysis of the CST6 proximal promoter and exon 1 in representative primary breast tumors and lymph node metastases. (A) The distribution of CpG dinucleotides proximal to the transcription start site in the promoter (0 to −1400 nucleotides) and exon 1 (0 to +294 nucleotides) of CST6 are depicted schematically (vertical lines indicate the relative position of individual CpG dinucleotides). Methylation analysis was performed on a region of the promoter spanning from −118 to +242 (indicated by a solid horizontal line), which contains 46 CpG dinucleotides and encompasses a large CpG island. (B) All clones analyzed for methylation of the CST6 promoter and exon 1 (46 CpGs) are shown for representative primary breast tumor and lymph node metastases examples. Black circles correspond to methylated CpGs and open circles correspond to unmethylated CpGs. TMI values for the entire promoter/exon 1 region (46 CpGs) are given for each primary breast tumor and lymph node metastases. NB1, P4, and P3 express cystatin M, while LNM1 and LNM5 lack cystatin M protein expression.
The majority (7/10, 70%) of metastatic lesions that are negative for cystatin M expression exhibit hypermethylation of the CST6 promoter/exon 1 region, with TMI values ranging from 11% to 46% (average TMI 17 ± 4%). The TMIs for cystatin M-negative lymph node metastases were found to be statistically increased relative to that determined for normal breast (P<0.0001). Fig. 4B shows representative methylation analyses. LNM1 and LNM5 were methylated at 17/46 (37%) and 38/46 (83%) CpGs, respectively, resulting in TMIs of 37% and 46% (Fig. 4B, LNM1 and LNM5). P4N1 and LNM2 were 100% or intermediately methylated at 35/46 (76%) and 38/46 (83%) CpGs respectively, with the remaining CpGs unmethylated, reflecting TMIs of 22% and 24%. Cystatin M negative nodes P2N1, LNM3, and LNM4 were intermediately methylated at 12/46 (26%), 34/46 (74%), and 18/46 (39%) CpGs, resulting in TMI values of 11%, 13%, and 13% respectively. There were 3 cystatin M-negative lymph nodes that displayed TMI values ranging from 1% to 3%. Two lymph node metastases were positive for cystatin M and exhibit low levels of methylation. In lymph node P1N1, 23/46 (50%) CpGs were methylated at an intermediate level and 23/46 (50%) CpGs were unmethylated. In lymph node P3N1, 8/46 (17%) CpGs were methylated at an intermediate level, and 38/46 (83%) CpGs were unmethylated. In total, 8/12 (67%) metastatic lesions from 5/7 (71%) patients displayed CST6 promoter hypermethylation.
CST6 Gene Methylation Correlates with Loss of Cystatin M Expression in a Subset of Primary Breast Tumors and Lymph Node Metastases
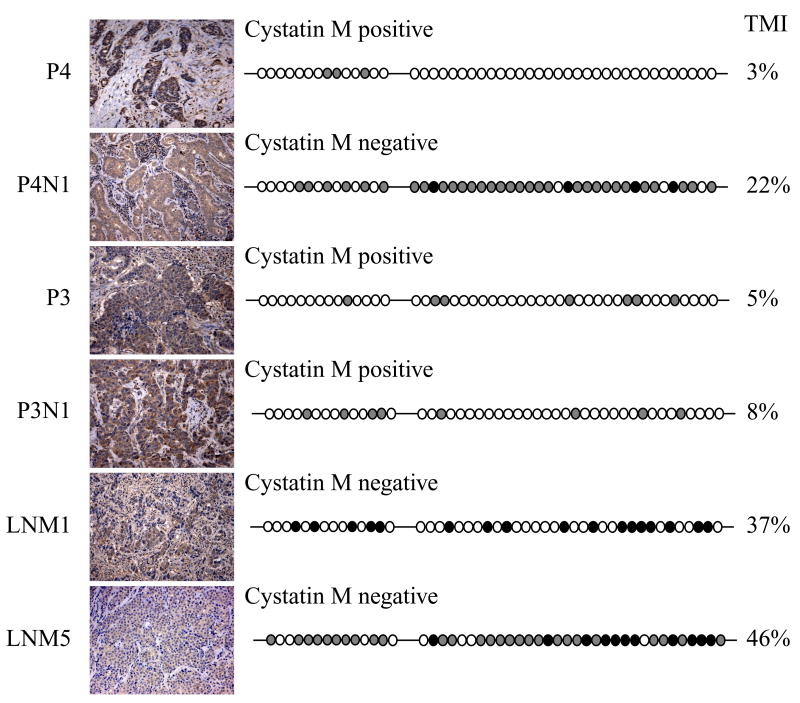
Cystatin M expression is associated with methylation status in the majority (12/23, 52%) of tumor tissues (Fig. 5). In most cases (4/7, 57%) cystatin M-positive tumors show a lack of CST6 promoter methylation (Fig. 5, P3, P3N1, and P4). In contrast, 3/7 (43%) cystatin M-positive tumors exhibit CST6 hypermethylation. This result suggests the existence of other epigenetic or genetic mechanisms that can bypass promoter hypermethylation. 8/16 (50%) cystatin M-negative tumors exhibit CST6 promoter hypermethylation with TMI values ranging from 11% to 46% (Fig. 5, P4N1, LNM1, and LNM5). These include one primary breast tumor and 7 lymph node metastases that lack expression of cystatin M. The remaining 8/16 (50%) cystatin M-negative tumors show very low levels of CST6 promoter methylation (TMI values ranging from 0% to 3%), including 5 primary tumors and 3 metastatic lesions. Overall, the extent of CST6 methylation corresponds with the expression of cystatin M expression in the majority (52%) of breast neoplasms, suggesting that methylation-dependent silencing of CST6 may represent an important mechanism for loss of cystatin M in a subset of breast neoplasms.
Fig. 5.
Correlation analysis of cystatin M expression and CST6 methylation status in primary breast tumors and lymph node metastases. Panels show cystatin M immunostaining (on left) and a summary of the methylation analysis of the CST6 promoter/exon 1 (46 CpGs) is show on the right. Black circles correspond to fully (100%) methylated CpGs, gray circles correspond to CpGs with intermediate methylation, and open circles correspond to unmethylated CpGs. TMI values for the entire promoter/exon 1 region (46 CpGs) are given for each tissue sample. P4 and P3 primary breast tumors, and lymph node metastasis P3N1 express cystatin M. Metastatic lesions P4N1, LNM1, and LNM5 show reduced expression of cystatin M. (Original objective lens magnification 10x).
Discussion
Cystatin M was originally described as exhibiting diminished expression in metastatic breast cancer, suggesting a role in suppression of the invasive/metastatic phenotype (Sotiropoulou et al., 1997). When exogenously expressed in human MDA-MB-435S breast cancer cells, CST6 significantly alters the neoplastic phenotype in vitro, resulting in diminished cell proliferation, loss of cell migration, inhibition of Matrigel invasion, and reduced endothelial cell adhesion (Shridhar et al., 2004). Furthermore, expression of CST6 in MDA-MB-435S breast cancer cells delays tumorigenesis by transplanted cells and suppresses spontaneous formation of liver and lung metastases (Zhang et al., 2004). More recently, it has been shown that CST6 is epigenetically regulated by DNA methylation-dependent silencing in breast cancer cell lines (Ai et al., 2006; Rivenbark et al., 2006a; Schagdarsurengin et al., 2006; Shridhar et al., 2004) and primary invasive ductal carcinomas (Ai et al., 2006; Schagdarsurengin et al., 2006). Ai et al. showed that 12/20 (60%) primary breast tumors exhibit CST6 promoter hypermethylation, and microdissection of individual cells from select tumors revealed that methylation occurs in both DCIS and IDC cells (Ai et al., 2006). In a similar study, Schagdarsurengin et al. showed that 24/40 (60%) breast carcinomas exhibited CST6 promoter hypermethylation, and that estrogen-receptor positive tumors were more frequently methylated than estrogen-receptor negative tumors (Schagdarsurengin et al., 2006). While CST6 is suggested to be epigenetically regulated through DNA methylation-dependent mechanisms in breast cancer cell lines and primary breast tumors that lack cystatin M protein expression, tumor metastases have not been examined for cystatin M expression or methylation status. Given a putative role for CST6 in suppression of tumor invasion and metastasis, loss of cystatin M expression may be one mechanism that enables tumor cells to spread from the primary site and invade adjacent tissues (or distant sites) during breast cancer progression. Furthermore, evidence from breast cancer cell lines suggests that CST6 promoter hypermethylation leading to gene silencing may represent one major mechanism for loss of cystatin M in breast cancer. CST6 is located in the chromosomal region 11q13, which is subject to amplification or loss of heterozygosity in several cancers (Cromer et al., 2004; Keppler, 2006; Srivatsan et al., 2002). Previously, we reported that the majority of CST6-negative breast cancer cell lines were originally established from metastatic lesions (pleural effusions) rather than primary breast tumors and that CST6-positive breast cancer cell line (BT-20) was derived from a primary breast carcinoma (Rivenbark et al., 2006a). These observations argue that the loss of CST6 expression is strongly associated with the invasive/metastatic phenotype of the breast cancer cell line and that CST6 promoter hypermethylation may be frequently involved in gene silencing/loss. In the current study, we present evidence that metastatic breast cancers exhibit lower levels of cystatin M protein expression and increased CST6 promoter hypermethylation compared to primary breast tumors.
The differential CpG island methylation of CST6 between primary breast tumors and lymph node metastases indicates that certain individual methylation events occur during or following stromal invasion and tumor spread. We envision that there is a succession of methylation events that lead to CST6 gene silencing in metastatic breast cancer: (i) individual CpG dinucleotides within the promoter region are preferentially methylated resulting in decreased expression of cystatin M in DCIS and/or primary breast carcinomas, (ii) methylation spreads throughout the CpG island during surrounding stromal invasion of tumor cells and metastasis to the regional lymph nodes resulting in a complete loss of cystatin M protein expression, and (iii) chromatin remodeling occurs resulting in the stable silencing of CST6. However, breast tumors that exhibit silencing of cystatin M but lack DNA methylation could achieve this silencing through histone deacetylation or through a putative transcription repressor binding to the promoter regulatory regions of CST6. Likewise, we have observed areas of tumor sections that show strong staining for cystatin M and areas that show weak staining. This finding may indicate that cystatin M silencing can be heterogeneous within a single breast tumor and can reflect different levels of DNA methylation.
In summary, 56% primary breast tumors and 85% lymph node metastases expressed significantly lower levels of cystatin M protein compared to normal breast tissue. CST6 promoter hypermethylation was seen in 48% of neoplastic lesions, and the extent of CST6 methylation correlates with loss of protein expression. In addition, lymph node metastases exhibited diminished levels of cystatin M when compared to matched primary breast tumors. These results suggest that methylation-dependent epigenetic silencing of CST6 represents an important mechanism for loss of CST6 during breast tumorigenesis and loss of CST6 expression is associated with the invasive/metastatic potential of breast cancer cells.
Acknowledgments
Grant support: This work was supported by grants from the Susan G. Komen Breast Cancer Foundation (BCTR0100575), the National Cancer Institute (NIH grant CA78343), and Sigma Xi, The Scientic Research Society Grand-In-Aid of Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ai L, et al. Epigenetic Silencing of the Tumor Suppressor Cystatin M Occurs during Breast Cancer Progression. Cancer Res. 2006;66:7899–7909. doi: 10.1158/0008-5472.CAN-06-0576. [DOI] [PubMed] [Google Scholar]
- Buck M, et al. Degradation of extracellular-matrix proteins by human cathepsin B from normal and tumour tissues. Biochem J. 1992;282:273–278. doi: 10.1042/bj2820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer A, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–98. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- Deng G, et al. Promoter methylation inhibits APC gene expression by causing changes in chromatin conformation and interfering with the binding of transcription factor CCAAT-binding factor. Cancer Res. 2004;64:2692–8. doi: 10.1158/0008-5472.can-03-3000. [DOI] [PubMed] [Google Scholar]
- Douglas DB, et al. Hypermethylation of a small CpGuanine-rich region correlates with loss of activator protein-2alpha expression during progression of breast cancer. Cancer Res. 2004;64:1611–20. doi: 10.1158/0008-5472.can-0318-2. [DOI] [PubMed] [Google Scholar]
- Frosch B, et al. Molecular regulation, membrane association and secretion of tumor cathepsin B. Apmis. 1999;107:28–37. doi: 10.1111/j.1699-0463.1999.tb01523.x. [DOI] [PubMed] [Google Scholar]
- Kane S, Gottesman M. The role of cathepsin L in malignant transfromation. Semin Cancer Biol. 1990;1:127–136. [PubMed] [Google Scholar]
- Keppler D. Towards novel anti-cancer strategies based on cystatin function. Cancer Lett. 2006;235:159–76. doi: 10.1016/j.canlet.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Kim TY, et al. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- Kos J, et al. Cysteine proteinases and their inhibitors in extracellular fluids: markers for diagnosis and prognosis in cancer. Int J Biol Markers. 2000;15:84–9. doi: 10.1177/172460080001500116. [DOI] [PubMed] [Google Scholar]
- Lah T, Kos J. Cysteine proteinases in cancer progression and their clinical relevance for prognosis. Biol Chem. 1998;379:125–130. [PubMed] [Google Scholar]
- Lau R, et al. Low levels of cell cycle inhibitor p27kip1 combined with high levels of Ki-67 predict shortened disease-free survival in T1 and T2 invasive breast carcinomas. Int J Oncol. 2001;18:17–23. doi: 10.3892/ijo.18.1.17. [DOI] [PubMed] [Google Scholar]
- Maciewicz RA, et al. Susceptibility of the cartilage collagens types II, IX and XI to degradation by the cysteine proteinases, cathepsins B and L. FEBS Lett. 1990;269:189–93. doi: 10.1016/0014-5793(90)81151-d. [DOI] [PubMed] [Google Scholar]
- Mai J, et al. Degradation of extracellular matrix protein tenascin-C by cathepsin B: an interaction involved in the progression of gliomas. Biol Chem. 2002;383:1407–1413. doi: 10.1515/BC.2002.159. [DOI] [PubMed] [Google Scholar]
- Mirza AN, et al. Prognostic factors in node-negative breast cancer: a review of studies with sample size more than 200 and follow-up more than 5 years. Ann Surg. 2002;235:10–26. doi: 10.1097/00000658-200201000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller V, et al. Sentinel Lymph Node Dissection and Micrometastasis Detection in Bone Marrow and Lymph Nodes. In: Ross JS, Hortobagyi GN, editors. Molecular Oncology of Breast Cancer. Jones and Bartlett; Boston: 2005. pp. 117–127. [Google Scholar]
- Ni J, et al. Cystatin E is a novel human cysteine proteinase inhibitor with structural resemblance to family 2 cystatins. J Biol Chem. 1997;272:10853–8. doi: 10.1074/jbc.272.16.10853. [DOI] [PubMed] [Google Scholar]
- Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–9. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Reed W, et al. The prognostic value of p53 and c-erb B-2 immunostaining is overrated for patients with lymph node negative breast carcinoma: a multivariate analysis of prognostic factors in 613 patients with a follow-up of 14–30 years. Cancer. 2000;88:804–13. doi: 10.1002/(sici)1097-0142(20000215)88:4<804::aid-cncr11>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Rivenbark AG, et al. DNA methylation-dependent silencing of CST6 in human breast cancer cell lines. Lab Invest. 2006a;86:1233–1242. doi: 10.1038/labinvest.3700485. [DOI] [PubMed] [Google Scholar]
- Rivenbark AG, et al. DNA methylation-dependent epigenetic regulation of gene expression in MCF-7 breast cancer cells. Epigenetics. 2006b;1:32–44. doi: 10.4161/epi.1.1.2358. [DOI] [PubMed] [Google Scholar]
- Roshy S, et al. Pericellular cathepsin B and malignant progression. Cancer Metastasis Rev. 2003;22:271–286. doi: 10.1023/a:1023007717757. [DOI] [PubMed] [Google Scholar]
- Schagdarsurengin U, et al. Frequent epigenetic inactivation of cystatin M in breast carcinoma. Oncogene. 2006 doi: 10.1038/sj.onc.1210107. [DOI] [PubMed] [Google Scholar]
- Shridhar R, et al. Cystatin M suppresses the malignant phenotype of human MDA-MB-435S cells. Oncogene. 2004;23:2206–15. doi: 10.1038/sj.onc.1207340. [DOI] [PubMed] [Google Scholar]
- Simpson PT, et al. Molecular evolution of breast cancer. J Pathol. 2005;205:248–54. doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]
- Slone BF, et al. Lysosomal cathpsin B: correlation with metastatic potential. Science. 1981;212:1151–1153. doi: 10.1126/science.7233209. [DOI] [PubMed] [Google Scholar]
- Song J, et al. The candidate tumor suppressor CST6 alters the gene expression profile of human breast carcinoma cells: down-regulation of the potent mitogenic, motogenic, and angiogenic factor autotaxin. Biochem and Biophy Res Comm. 2006;340:175–182. doi: 10.1016/j.bbrc.2005.11.171. [DOI] [PubMed] [Google Scholar]
- Sotiropoulou G, et al. Identification, cloning, and characterization of cystatin M, a novel cysteine proteinase inhibitor, down-regulated in breast cancer. J Biol Chem. 1997;272:903–10. doi: 10.1074/jbc.272.2.903. [DOI] [PubMed] [Google Scholar]
- Srivatsan ES, et al. Localization of deletion to a 300 Kb interval of chromosome 11q13 in cervical cancer. Oncogene. 2002;21:5631–42. doi: 10.1038/sj.onc.1205698. [DOI] [PubMed] [Google Scholar]
- Symmans WF. Histopathology of Breast Cancer: Correlation with Molecular Markers. In: Ross JS, Hortobagyi GN, editors. Molecular Oncology of Breast Cancer. Jones and Bartlett; Boston: 2005. pp. 106–116. [Google Scholar]
- Vigneswaran N, et al. Silencing of cystatin M in metastatic oral cancer cell line MDA-686Ln by siRNA increase cysteine proteinases and legumain activities, cell proliferation and in vitro invasion. Life Sciences. 2006;78:898–907. doi: 10.1016/j.lfs.2005.05.096. [DOI] [PubMed] [Google Scholar]
- Zeeuwen PL. Epidermal differentiation: the role of proteases and their inhibitors. Eur J Cell Biol. 2004;83:761–73. doi: 10.1078/0171-9335-00388. [DOI] [PubMed] [Google Scholar]
- Zeeuwen PL, et al. Cystatin M/E expression in inflammatory and neoplastic skin disorders. Br J Dermatol. 2002;147:87–94. doi: 10.1046/j.1365-2133.2002.04785.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. Cystatin m: a novel candidate tumor suppressor gene for breast cancer. Cancer Res. 2004;64:6957–64. doi: 10.1158/0008-5472.CAN-04-0819. [DOI] [PubMed] [Google Scholar]
- Zhong S, et al. Pharmacologic inhibition of epigenetic modifications, coupled with gene expression profiling, reveals novel targets of aberrant DNA methylation and histone deacetylation in lung cancer. Oncogene. 2006 doi: 10.1038/sj.onc.1210041. [DOI] [PubMed] [Google Scholar]