Abstract
Background
Treatment of many inherited liver enzyme deficiencies requires the removal of toxic intermediate metabolites from the blood of affected individuals. We propose that circulating toxins can be adequately cleared and disease phenotype influenced by enzyme expressed in tissues other than the liver, such as bone marrow. Our specific hypothesis was that phenylalanine hydroxylase (PAH) expressed in bone marrow would lower blood phenylalanine levels in hyperphenylalaninemic Pahenu2 mice, a model of human phenylketonuria (PKU).
Methods
Germline-modified marrow PAH-expressing mice were developed using a transgene that contained the mouse liver PAH cDNA under the transcriptional control of a human β-globin promoter. Marrow PAH-expressing mice were bred to Pahenu2 mice to generate progeny that lacked liver PAH activity but expressed PAH in bone marrow.
Results
Marrow PAH expression did not affect the health, function, or reproductive capacity of transgenic animals. Hyperphenylalaninemia persisted in transgenic Pahenu2 homozygous mice despite PAH activity in marrow lysates, and was not altered following supplementation with tetrahydrobiopterin (BH4), a required cofactor for PAH. PAH activity measured in intact marrow cells was significantly lower than in marrow lysates; no such difference was measured in isolated hepatocytes vs. liver homogenate.
Conclusions
Marrow PAH expression did not correct hyperphenylalaninemia in Pahenu2 mice. Phenylalanine clearance may have been limited by the natural perfusion rate of the marrow compartment, by insufficient PAH expression in marrow, or by other cellular factors affecting phenylalanine metabolism in intact marrow cells. Differences in PAH activity measured in intact marrow cells vs. cell lysates suggest that hepatocytes and PAH-expressing marrow cells are fundamentally different in their ability to metabolize phenylalanine. The efficacy of bone-marrow-directed gene therapy as a metabolic sink in the treatment of phenylketonuria may be limited, although further experiments with greater marrow PAH expression levels will be necessary to definitively prove this conclusion.
Keywords: inborn errors of metabolism, hyperphenylalaninemia, phenylketonuria, bone marrow gene therapy, β-globin gene, phenylalanine hydroxylase
Introduction
Many inborn errors of metabolism (IEM) are caused by deficiencies of intermediate enzymes in liver. For example, inherited deficiency of liver phenylalanine hydroxylase (PAH, EC 1.14.16.1) causes phenylketonuria (PKU), one of the most common IEM in humans [1]. Restoration of enzyme activity through liver-directed gene transfer is an obvious potential approach for treatment of these disorders. However, in select IEM, the pathophysiology of the disease is actually due to circulating toxins that accumulate in the context of liver enzyme deficiency, and the enzyme deficiency itself has no direct pathologic effect upon liver. We propose that ectopic expression of enzyme in tissues other than liver (so-called heterologous gene therapy) could effectively clear circulating toxins and favorably influence disease phenotype. In this study, we examined the efficacy of erythrogenic bone marrow as a potential platform for heterologous gene therapy of IEM using the Pahenu2 mouse, a murine model of human PKU. Pahenu2 homozygous mice carry a missense mutation in the PAH gene that results in complete liver PAH deficiency and hyperphenylalaninemia [2]. Rather than rely upon direct gene transfer into bone marrow of hyperphenylalaninemic mice, we chose to employ germline modification techniques to induce bone marrow PAH activity in transgenic mice and thereby thoroughly examine the potential efficacy of bone-marrow-directed gene therapy for the treatment of murine PKU.
Materials and methods
Characterization of the μLCR promoter
We chose to induce erythrogenic marrow-specific PAH expression using a modified 2.8 kb gene fragment from the human β-globin locus control region (LCR), kindly provided by Dr. George Stamatoyannopoulos (Seattle, WA, USA). This μLCR sequence contained the four expression-enhancing DNase I hypersensitive (HS) sites. This μLCR sequence has previously been shown by other investigators to direct erythrogenic marrow expression in transgenic mice [3]. The μLCR sequence was supplied as an insert in pBluescript (SK-) (Stratagene, La Jolla, CA, USA) in the Hind III site (plasmid named pμLCR). In order to evaluate the ability of the μLCR promoter to direct gene expression from a recombinant vector in erythrogenic cells, we constructed a firefly luciferase cDNA-containing recombinant plasmid (pμLCRGL3) by inserting a 3.2 kb Hind III fragment containing the μLCR promoter from pμLCR into a commercially available reporter plasmid, pGL3 Enhancer (Promega Corp., Madison, WI, USA). Luciferase expression from the pμLCRGL3 plasmid was tested following transient transfection into murine erythroleukemia (MEL) cells using liposomes. Luciferase expression from pμLCRGL3 in MEL cells was compared with that from a commercially available control plasmid (pCILuc) containing the CMV early enhancer/promoter.
Liposome-mediated DNA transfection into MEL cells
pμLCRGL3 plasmid DNA was transfected into MEL cells in tissue culture using TransIt LT-1 transfection reagent (Mirus Corp., Madison, WI, USA) according to the manufacturer's instructions. In brief, DNA and LT-1 reagent were mixed in a 1 : 3 ratio by weight and diluted in 1 ml tissue culture media (RPMI + 10% fetal bovine serum) containing 106 MEL cells in a T-25 flask. The cells were incubated at 37 °C for 2 h and 2 ml fresh medium were then added. 20-μl and 100-μl aliquots of cell culture suspension were removed daily for cell count and luciferase assay, respectively.
Luciferase assay
Transiently transfected MEL cells in suspension were briefly centrifuged at 10 000 g in an Eppendorf tube to pellet the cells. The media was removed and the cells were washed once with 1 ml phosphate-buffered saline (PBS), pH 7.4. The cell pellet was resuspended in 100 μl lysis buffer (0.1 M potassium phosphate buffer, pH 7.8, 1 mM dithiothreitol, 0.1% Triton X-100) and lysed by three freeze-thaw cycles in a dry ice/ethanol bath. Following centrifugation, luciferase assay was performed on 20 μl of cell lysate using standard procedures [4] using a Luminescence laboratory luminometer. Data were expressed as relative light units (RLU).
Construction of LCRβgmPAH transgene and development of LCRPAH mice with bone marrow PAH expression
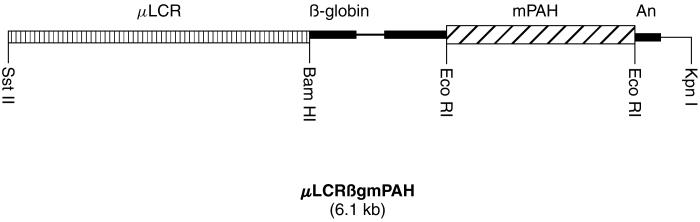
Once we had confirmed the ability of the μLCR promoter to direct gene expression in vitro, we began construction of the LCRβgmPAH transgene for the development of germline-modified mice with PAH expression in erythrogenic bone marrow (hereafter designated LCRPAH mice). The LCRβgmPAH transgene (Figure 1) contained a mouse liver PAH (mPAH) cDNA [5] inserted into an Eco RI site within the second exon of the rabbit genomic β-globin gene. Construction of a pBluescript (KS)-based vector (pBlueβgmPAH) containing the mouse liver PAH (mPAH) cDNA inserted in the rabbit β-globin gene has been described previously [6]. We created an Sst II site 5′ to Cla I in the pμLCR plasmid using PCR-based site-directed mutagenesis. The primers LCR1 (5′-GGGCCCCCCCTCGAGGTCCGCGG-3′) and LCR2 (5′-CCCTCCTCCCAGGTCCACGTGC-3′) were employed to amplify a 336 bp fragment of pμLCR. LCR1 contained a two base-pair mismatch that created the Sst II site in the final PCR product. The mutant PCR product was inserted back into pμLCR via the Xho I and Pml I sites. Then, a 3.2 kb Sst II-Bam HI fragment was isolated and subcloned into pBlueβgmPAH to create the final plasmid construct. The full 6.1 kb LCRβgmPAH transgene was cleaved from the plasmid vector through Sst II/Kpn I digestion and purified by agarose gel electrophoresis, reverse-phase column chromatography (Elutip™-d column, Schleicher and Schuell), and ethanol precipitation. The purified DNA was resuspended in 10 mM Tris-HCl, pH 7.4, 0.25 mM EDTA buffer for microinjection into nuclei of FVB/n embryos according to standard techniques for development of transgenic mice. Genomic DNA was analyzed for the presence of the LCRβgmPAH transgene by Eco RI or Bam HI digestion followed by Southern blotting. A Bam HI/Eco RI fragment from the rabbit β-globin gene was 32P-labeled by the random priming method and used as the probe.
Figure 1.
μLCRβgmPAH transgene. Expression of the mouse PAH cDNA is directed by the locus control region (μLCR) of the human β-globin gene [3]. An intronic sequence from the rabbit β-globin gene has been added to enhance cDNA expression in a transgenic mouse
Breeding of LCRPAH mice to Pahenu2/Pahenu2 mice
Female LCRPAH mice were bred to Pahenu2/Pahenu2 males. By Mendelian principles, all progeny were Pahenu2 heterozygotes and 50% inherited the LCRβgmPAH transgene. Two different breeding schemes were then followed. First, female LCRβgmPAH-positive, Pahenu2 heterozygous mice were bred to Pahenu2/Pahenu2 males to produce progeny (hereafter designated Tg/Pahenu2 mice) which were both homozygous for the Pahenu2 mutation and carried the LCRβgmPAH transgene in single copy. Alternatively, LCRβgmPAH-positive, Pahenu2 heterozygous siblings were bred together. In the F2 generation, 25% of the progeny are Pahenu2/Pahenu2. Of these, one-half carried a single copy of the LCRβgmPAH transgene and one-quarter inherited two doses of the transgene.
Detection of the Pahenu2 mutation by PCR analysis
The presence of the nt.788T→C mutation in the seventh exon of the murine PAH gene eliminates a Bbs I restriction site and creates a Bsm AI recognition site. A PCR-based restriction fragment length polymorphism method was used to detect the presence of the Pahenu2 mutation as previously described [6].
Phenotypic characterization of bone marrow PAH-expressing mice
Serum phenylalanine concentrations were measured in 25 μl serum using a modification of a fluorometric procedure [7].
PAH activity was measured in whole cells or tissue homogenates using a radiochemical technique modified from Ledley et al. [8], as previously described [6]. For measurement of bone marrow PAH activity, marrow was removed from dissected femurs by flushing the marrow compartment with 0.5 ml normal saline or Hank's balanced salt solution. Marrow samples from the two femurs of one mouse were combined for PAH assay. An aliquot of bone marrow suspension was removed for cell count by trypan blue exclusion in a hemocytometer. For measurement of PAH activity in marrow cell lysates, the marrow cell pellet was collected by centrifugation, resuspended in 100 μl homogenization buffer (1.15 M KCl, 0.1% 2-mercaptoethanol), lysed by three cycles of freeze-thawing in a dry ice/ethanol bath, then clarified by centrifugation. For the PAH reaction, 71.5 μl of marrow lysate were mixed with 10 μl 2 M potassium phosphate buffer, pH 6.8, and 10 μl 1.5 M KCl. For measurement of PAH activity in intact marrow cells, 5 × 106 marrow cells were pelleted by centrifugation and resuspended in 85 μl RPMI medium, pH 7.4. The following reagents were then added sequentially to each lysate or cell suspension: 2 μl 25 000 U/ml catalase, 1 μl 60 mM L-phenylalanine, 6 μlL-[U-14C]phenylalanine (0.3 μCi; Amersham). Following incubation for 5 min at room temperature, 2 μL of 0.1 M dithiothreitol (DTT) were added and PAH reactions initiated by adding 2 μl of 4.5 mM 6-methyltetrahydropterin (Calbiochem). Reactions were carried out at room temperature for 1 h then halted by boiling. Radiolabeled tyrosine produced in the reaction was separated from phenylalanine by thin-layer chromatography, then imaged and quantitated using a Biorad FX molecular imager and Quantity One densitometry software. Control reactions included buffer alone, LCRPAH mouse liver homogenate or intact LCRPAH hepatocytes isolated by in situ collagenase perfusion [9], and duplicate reactions lacking 6-methyltetrahydropterin cofactor. For cell lysates and liver homogenate, PAH activity was corrected for total protein as measured using a bicinchonic acid procedure (Microprotein Assay, Pierce, Rockford, IL, USA). PAH activity in intact cells was corrected for the number of cells in the reaction.
Tetrahydrobiopterin administration
6(R)-L-erythro-5,6,7,8-tetrahydrobiopterin (BHy) (Schircks Laboratories, Jona, Switzerland) was dissolved in 1% ascorbic acid, pH 7.0, to 100 mM and administered to mice by intraperitoneal injection using a 27-gauge needle on a 1-ml disposable syringe.
Tetrahydrobiopterin uptake into tissues
BH4 (0.1 μmol BH4/gm body weight, total volume = 200–300 μl) in 1% ascorbic acid was injected intravenously via the tail vein of sedated LCRPAH mice. At various time points following BH4 injection, the mice were euthanized by exsanguination via cardiac puncture, perfused via the left ventricle with 0.9% saline and tissue samples were collected. Tissues were also collected from mice injected with only 1% ascorbic acid. All solid tissue samples were weighed and homogenized in five volumes of 50 mM Tris/1 mM EDTA/1 mM DTT, pH 7.4. Bone marrow was collected as described above by perfusing the femur marrow cavity with 0.9% saline. Following centrifugation of the perfusate, the cell pellet was lysed in 100 μl 50 mM Tris/1 mM EDTA/1 mM DTT, pH 7.4. Supernatants from all tissues were removed and frozen at −20 °C until BH4 analysis. Total biopterin was measured by HPLC using established methods [10] in the laboratory of Dr. Sheldon Milstien, National Institute of Mental Health, Bethesda, MD, USA.
Results
Evaluation of μLCR-driven in vitro expression
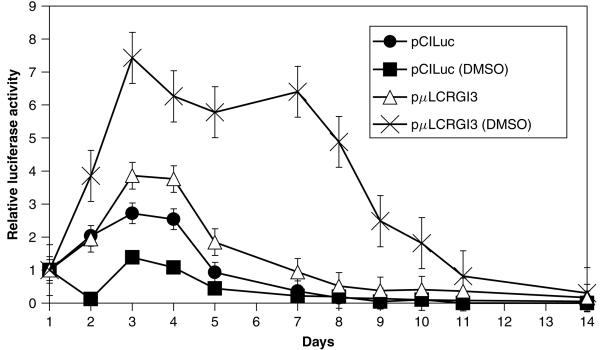
To evaluate expression from the μLCR sequence in our hands, we measured luciferase activity in MEL cells following liposome-mediated transient transfection of pμLCRGL3 vs. transfection with the control plasmid pCILuc. After transfection, MEL cultures were fed either with standard media alone or media containing 2% dimethyl sulfoxide (DMSO) to induce erythroid differentiation. The cultures were sampled daily thereafter for measurement of luciferase activity. These data are presented in Figure 2 as the amount of luciferase activity relative to luciferase expression on the first day post-transfection. Cultures receiving pCILuc produced higher absolute levels of luciferase activity, often approaching 1000-fold more luciferase light units per sample than cultures receiving pμLCRGL3. However, luciferase expression was sustained for a longer period in cells that received pμLCRGL3 plasmid. In each culture, regardless of the specific plasmid introduced into the cells or whether the culture media contained DMSO, absolute luciferase expression peaked on the third day following transfection. However, luciferase expression from pCILuc diminished rapidly thereafter, falling below the day 1 activity level by 5 days post-transfection. The presence of DMSO in the media had no effect on the duration of luciferase expression from pCILuc although peak absolute levels of luciferase activity were modestly lower in differentiated MEL cells. Luciferase expression from pμLCRGL3 in undifferentiated MEL cells lasted slightly longer, falling below the day 1 activity level on day 7 post-transfection. Most importantly, luciferase expression in differentiated MEL cells that had received pμLCRGL3 was sustained at high levels for up to 10 days post-transfection. This experiment demonstrates the μLCR promoter sequence in our plasmids was capable of directing sustained expression in differentiated erythroid cells.
Figure 2.
In vitro evaluation of μLCR-directed expression in MEL cells. Luciferase expression vectors containing either the CMV early promoter region (pCILuc) or the β-globin μLCR promoter (pμLCRGl3) were transfected into 106 MEL cells in 25-cm2 flasks using Trans-It LT-1 transfection reagent. Following transfection, 2% DMSO was added to the media of half the flasks to induce erythroid differentiation. Aliquots containing approximately 105 cells were sampled 24 h post-transfection then daily thereafter for measurement of luciferase activity. The data are presented as the ratio of measured luciferase activity relative to expression on day 1. In MEL cells, the μLCR promoter directs longer lasting and more robust expression than the CMV promoter especially under conditions stimulating erythroid differentiation
Development of bone marrow PAH-expressing mice by germline modification
Once we had verified that the μLCR promoter in our hands was active in erythrogenic cells, we designed and constructed a PAH-expressing transgene (μLCRβgmPAH) that contained a mouse PAH cDNA driven by the μLCR promoter (Figure 1). The intronic sequence from the rabbit β-globin gene was included to enhance transgene expression in germline-modified mice [11]. The first translation start site (ATG) is located in the mouse PAH cDNA 53 bp downstream from the 5′ EcoRI site, and a native polyadenylation signal is present in the mouse PAH cDNA downstream from the open reading frame. Transcription of the transgene yields a putative 2.9 kb pre-mRNA that is reduced to 2.3 kb after removal of the β-globin intronic sequence by splicing. Translation yields the 454 amino acid murine PAH monomer. No rabbit β-globin amino acid sequences are present in the final polypeptide. Transient transfection of the final transgene construct into MEL cells yielded negligible PAH activity (data not shown). However, in our experience, in vitro PAH expression from transgenes with tissue-specific promoters does not necessarily correlate with tissue PAH activity in germline-modified mice [6].
The μLCRβgmPAH transgene was cleaved and purified from its plasmid vector and introduced into single cell mouse embryos by microinjection using standard techniques [12]. Injected embryos were implanted into pseudopregnant foster dams and carried to term. Genomic DNA from the progeny was screened for the presence of the transgene with Southern blotting. Out of 46 live-born mice, 5 animals were found to carry the μLCRβgmPAH transgene. These five animals were bred to nontransgenic siblings and four of the five animals successfully produced transgenic offspring and led to the development of a founder line. One of the four founder lines carried approximately 2–3 copies of the transgene in a tandem arrangement and expressed higher levels of PAH activity in bone marrow than the other founder lines. This founder line (designated LCRPAH) was utilized in the remaining experiments.
Substantial PAH activity was detected in bone marrow lysates and spleen homogenates from LCRPAH heterozygous mice (Table 1). PAH activity was normal in liver homogenates and was detected in pancreas and kidney (data not shown) where it is also expressed in wild-type mice. PAH activity was undetectable in LCRPAH plasma and unfortunately in lysates of peripheral erythrocytes. Additionally, PAH activity was not detected in homogenates of whole brain, cardiac or skeletal muscle. Transgene expression was restricted to hematopoietic tissues including bone marrow and spleen of LCRPAH mice.
Table 1.
Tissue phenylalanine hydroxylase (PAH) activity
| Specific activity (nmol tyrosine/mg protein/h) Tissue |
|||
|---|---|---|---|
| Genotype | Liver | Bone Marrow | Spleen |
| +/+ | 212 ± 55 n = 10 |
0.015 ± 0.02 n = 2 |
0.4 n = 1 |
|
Pahenu2/Pahenu2 n = 10 |
0.27 ± 0.11 | Not measured | Not measured |
| LCRPAH n = 5 |
87.9 ± 26.2 | 10.7 ± 1.35 | 17.4 ± 7.1 |
| Tg/Pahenu2 n = 6 |
0.66 ± 0.19 | 1.92 ± 0.55 | 7.75 ± 2.94 n = 3 |
LCRPAH – parent transgenic line, heterozygous for μLCRßgmPAH transgene.
Tg/Pahenu2 – heterozygous for μLCRßgmPAH transgene, homozygous for Pahenu2.
LCRPAH mice appeared healthy, active, exhibited normal weight gain, and normal reproductive behavior. Approximately 50% of the offspring from LCRPAH carrier mice were also transgenic indicating the absence of any negative selection against the transgene. Furthermore, animals homozygous for the transgene were generated by breeding male and female LCRPAH mice. The presence of a double dose of the transgene was evaluated by densitometric analysis of Southern blots and confirmed through further breeding experiments. All offspring of putative LCRPAH homozygous mice carried the transgene indicating that the parent was in fact homozygous for the transgene. The mating of two LCRPAH homozygous mice yielded normal litter sizes and only homozygous offspring. From that point forward, the LCRPAH transgenic line was maintained through mating of homozygous mice.
Bone marrow PAH-expressing LCRPAH-heterozygous mice were evaluated for abnormalities of erythropoiesis by examination of peripheral blood. Peripheral blood hematocrit and hemoglobin concentrations were similar in LCRPAH mice and their nontransgenic siblings. The histologic appearance of Wright stained peripheral blood smears was also normal in LCRPAH mice. PAH expression in erythrogenic bone marrow and spleen did not adversely affect erythropoiesis or the health or reproductive capabilities of LCRPAH mice.
Generation of bone marrow PAH-expressing mice that lack liver PAH activity
In order to test the hypothesis that bone marrow PAH expression could cure phenylketonuria, bone marrow PAH-expressing LCRPAH mice were bred to hyperphenylalaninemic Pahenu2 mice, a murine model of human phenylketonuria. In this breeding scheme, female LCRPAH/+ mice were mated to male Pahenu2/Pahenu2 to generate offspring that were heterozygous for both the LCRPAH transgene and the Pahenu2 mutation. Two different mating schemes were then followed to generate animals that both carried the μLCRβgmPAH transgene and were homozygous for the Pahenu2 mutation (Tg/Pahenu2). First, female LCR+, Pahenu2/+ mice were mated to male Pahenu2/Pahenu2 mice. One-quarter of the offspring from this mating were expected to both carry the μLCRβgmPAH transgene and be homozygous for the Pahenu2 mutation. Out of seventeen progeny from two litters, six mice were LCR+ and Pahenu2/Pahenu2, a result that is not significantly different from the expected number of 4.25 Tg/Pahenu2 mice (p = 0.8, by χ2 test). Again, this result confirms the lack of any negative selection pressure against LCR+, Pahenu2/Pahenu2 mice. In the second breeding approach, Tg/Pahenu2 mice were generated through mating of LCR+, Pahenu2/+ siblings. Only 3/16 of the progeny were expected to be Tg/Pahenu2, but this mating has the advantage of producing animals that are both Pahenu2/Pahenu2 and carry two copies of the μLCRβgmPAH transgene. These animals provided the opportunity to evaluate whether increasing the amount of PAH activity in bone marrow from a double dose of the transgene would have a further effect upon serum phenylalanine levels in Pahenu2/Pahenu2 mice. Between the two breeding schemes, a total of eighteen Tg/Pahenu2 mice were generated out of 91 total offspring.
PAH activity was detected in bone marrow lysates and spleen homogenates from Tg/Pahenu2 mice, while liver PAH activity was virtually absent (Table 1). Liver PAH activity in Tg/Pahenu2 mice was not significantly different from that of Pahenu2/Pahenu2 mice, confirming that Tg/Pahenu2 mice were in fact homozygous for the Pahenu2 mutation. Bone marrow and spleen PAH activities of Tg/Pahenu2 measured somewhat lower than the activities in marrow and spleen of the parent LCRPAH transgenic line. These differences may have been due to variability in the PAH assay as the measurements were performed immediately after euthanasia and tissue harvest from each mouse and therefore required multiple independent assays over several months. Repeat assays in bone marrow from an individual mouse were not possible as the entire lysate was consumed in the initial PAH assay. No change in the number of transgene tandem repeats was evident from densitometry of Southern blots on LCRPAH and Tg/Pahenu2 mice. Unfortunately, Tg/Pahenu2 mice remained hyperphenylalaninemic despite substantial bone marrow and spleen PAH activity (Table 2), and serum phenylalanine levels of Tg/Pahenu2 mice did not differ significantly from levels in Pahenu2/Pahenu2 animals.
Table 2.
Serum phenylalanine levels of Tg/Pahenu2 mice
| Genotype | Serum phenylalanine (μmol/l) Mean ± SD |
|---|---|
| +/+ (n = 10) |
138 ± 12 |
|
Pahenu2/Pahenu2 (n = 11) |
1574 ± 380 |
|
Tg/Pahenu2 (n = 13) |
1710 ± 164 |
Possible explanations for this result include (1) that bone marrow PAH activity was still insufficient to adequately clear phenylalanine from the circulation, (2) that cellular factors other than PAH activity (such as phenylalanine membrane transport) in erythrogenic marrow could limit phenylalanine clearance, (3) that PAH-expressing marrow cells contained insufficient BH4 to support physiologically relevant levels of phenylalanine hydroxylation, or (4) that phenylalanine clearance was limited by the rate of vascular flow and hence phenylalanine flux through the marrow compartment. We performed formal experiments to evaluate all these possibilities.
Phenylalanine clearance in bone marrow PAH-expressing mice was probably not limited by the amount of PAH activity in marrow lysates
Pahenu2/Pahenu2 animals that had inherited two copies of the LCRPAH transgene exhibited greater levels of bone marrow PAH activity than heterozygous mice. Two such mice had bone marrow PAH activity of 2.9 and 2.4 nmol tyrosine produced/mg protein/hour compared with a mean activity of 1.9 ± 0.5 in heterozygotes. For these measurements, we assayed PAH activity in approximately 3 × 105 marrow cells from an LCRPAH homozygous animal, and the assay yielded 10.3 nmol tyrosine produced per hour. Marrow isolated from both femurs, tibias and humeri of an adult mouse yields approximately 2 × 108 cells (Jane Barker, Jackson Laboratory, personal communication). Therefore, we estimate that the total phenylalanine hydroxylation capacity of the entire bone marrow from a transgene homozygous mouse was about 7 × 103 nmol tyrosine produced/h. The phenylalanine hydroxylation capacity of the entire liver of a wild-type C57Bl/6J mouse as measured in liver homogenate was 4.7 × 104 nmol tyrosine/h.
Phenylalanine clearance is limited in intact LCRPAH marrow cells
Phenylalanine hydroxylation was measured in intact marrow cells and isolated primary hepatocytes from homozygous LCRPAH mice (n = 6). The phenylalanine hydroxylation rate in intact LCRPAH marrow cells was 2.73 (±1.35) ×10−7 nmol tyrosine produced/h/cell (mean ± SE) compared with 2.09 (±0.676) ×10−4 nmol/h/cell in isolated LCRPAH hepatocytes. Increasing the concentration of the 6-methyltetrahydropterin cofactor 10-fold in the reaction did not increase tyrosine production from LCRPAH marrow cells (0.62 × 10−7 nmol/h/cell). No tyrosine production was detected in Pahenu2/Pahenu2 marrow cells. Assuming that an adult mouse liver contains approximately 1 × 108 hepatocytes and that 2 × 108 cells are present in the marrow compartment, then the phenylalanine hydroxylation capacity as measured in intact cells of the entire marrow of the LCRPAH mouse was only 54.6 nmol tyrosine produced/h compared with 2.09 × 104 nmol/h from the entire liver.
Phenylalanine clearance in bone marrow PAH-expressing mice was not limited by BH4 supply
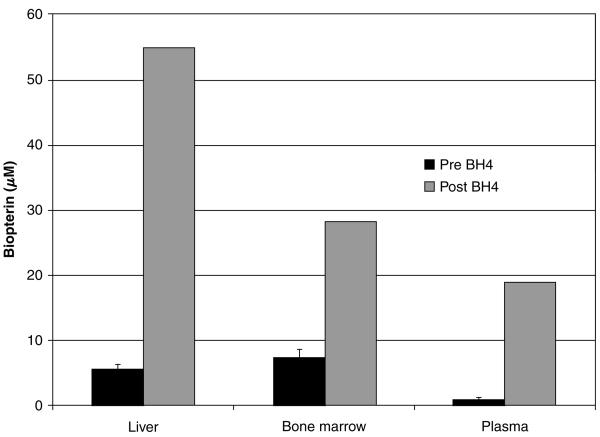
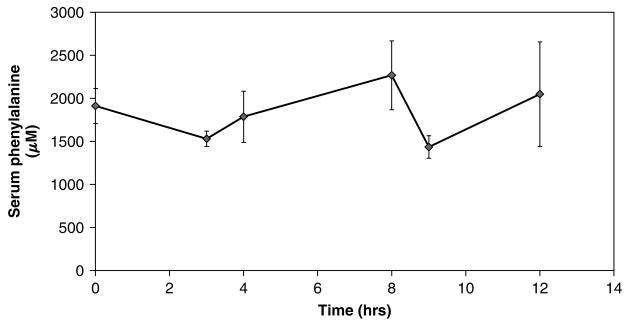
Previously published reports [10] suggest that bone marrow contains a significant amount of BH4 compared with other tissues and almost as much as liver. To confirm this finding, we measured BH4 content of bone marrow homogenate from LCRPAH mice using HPLC with the assistance of Dr. Sheldon Milstien, National Institute of Mental Health, Bethesda, MD, USA. Bone marrow homogenates from LCRPAH mice contained a substantial amount of BH4 that should have been sufficient to support physiologically meaningful phenylalanine hydroxylation (Figure 3). Regardless, we attempted to enhance bone-marrow-directed phenylalanine hydroxylation through administration of exogenous BH4. In a separate experiment, we verified that BH4 administered to LCRPAH mice via intravenous injection did in fact penetrate the marrow cells (Figure 3). BH4 was administered hourly to Tg/Pahenu2 mice via intraperitoneal injection. Serum phenylalanine levels of Tg/Pahenu2 mice did not change following BH4 administration (Figure 4) at least over the time course of this trial.
Figure 3.
Tissue biopterin content. Tissue total biopterin (μM) in LCRPAH mice before and 1 h after intravenous injection of BH4, 0.1 μmol/gm body weight in 1% ascorbic acid. Baseline tissue biopterin levels (Pre BH4) were measured in six mice while tissue biopterin following intravenous BH4 injection was evaluated in only a single animal. Data for Pre BH4 injection tissue biopterin is expressed as mean (μM) ±SE
Figure 4.
Hourly BH4 administration to Tg/Pahenu2 mice. BH4 in 1% ascorbic acid was administered hourly by intraperitoneal injection to Tg/Pahenu2 mice (n = 4–8 mice for each time point). Blood was sampled repetitively from the tail vein for measurement of serum phenylalanine level. Data are expressed as mean serum phenylalanine (μM) ±SE vs. time (hrs)
BH4 uptake into bone marrow is relatively slow compared with skeletal muscle
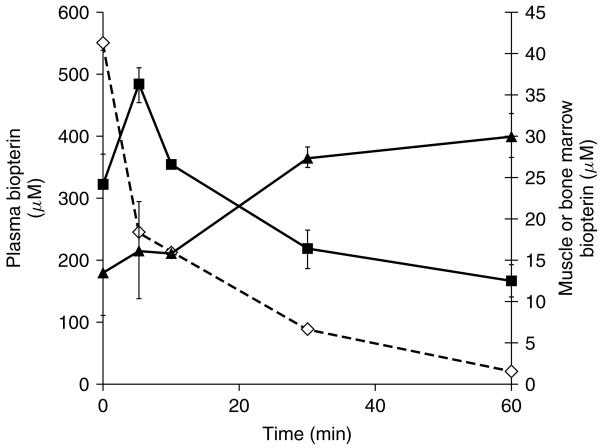
Following intravenous administration of BH4 to LCRPAH mice, we measured biopterin levels in several tissues at various time points after injection. BH4 was rapidly cleared from the plasma with a half-life of 12 min. Most of the injected BH4 is rapidly taken up by liver and kidney (data not shown), two organs that receive large blood flows. Although levels of BH4 in marrow increased to levels above that measured in skeletal muscle post-injection, the rate of BH4 uptake into marrow was slower than into muscle (Figure 5). Muscle BH4 levels peaked within 5 min post-injection and then decreased with a half-life of approximately 27 min. Marrow BH4 levels continued to increase for up to 30 min following injection and then remained relatively constant at least up to 60 min post-injection.
Figure 5.
Tissue BH4 concentration vs. time following IV BH4 injection. BH4 in 1% ascorbic acid was administered to LCRPAH mice by intravenous injection. Animals were euthanized and tissues harvested for BH4 determination at various time points following injection. Data are expressed as BH4 concentration (μM) vs. time (min). Plasma BH4 (-◇-) is plotted on left Y-axis, while muscle (-■-) and marrow (-▲-) are plotted on the right Y-axis
Discussion
Our previous work with PAH expression in muscle of hyperphenylalaninemic mice demonstrated the successful reduction of serum phenylalanine levels in the animals, but only if sufficient BH4 was supplied exogenously [6]. Muscle BH4 supply is the major limiting factor to the efficacy of muscle-directed gene therapy of PKU using PAH. Cultured keratinocytes have also been investigated as a platform for ectopic phenylalanine metabolism [13]; this system is also limited by the lack of native BH4 synthesis and requires the coexpression of both PAH and BH4 biosynthetic enzymes. Native BH4 content is significantly higher in bone marrow and in circulating erythrocytes than in muscle, approximating the levels found in liver. PAH has been expressed in circulating human T lymphocytes in vitro following recombinant retroviral vector administration [14], but the effectiveness of this approach as a metabolic sink for phenylalanine has not, to our knowledge, been demonstrated in vivo. We proposed that phenylalanine hydroxylation mediated by PAH activity in marrow or in peripheral red blood cells would not be limited by BH4 supply. Given the very large number of erythrocytes in peripheral blood, we predicted that if PAH expression in erythrogenic marrow had resulted in persistent PAH activity within circulating mature erythrocytes, then phenylalanine clearance rates in liver PAH-deficient mice should have improved dramatically. Our confidence that PAH activity would persist in circulating erythrocytes was however tempered from the outset by the knowledge that the developing reticulocyte employs a robust proteolytic system to generally remove most intracellular protein, save hemoglobin and a handful of other proteins, prior to emergence of the mature erythrocyte. As feared, no PAH activity was detected within circulating erythrocytes of Tg/Pahenu2 mice. Still, we estimated from measurements in tissue lysates that the amount of PAH activity expressed in Tg/Pahenu2 bone marrow and spleen should have been sufficient to effectively clear phenylalanine.
Unfortunately, marrow PAH expression had no phenotypic effect on hyperphenylalaninemic, liver PAH-deficient mice. Mice with a double dose of the μLCRβgmPAH transgene exhibited approximately double the amount of PAH activity in marrow lysate compared with merely heterozygous mice. In published gene transfer experiments utilizing PAH cDNA-containing recombinant adenovirus to treat Pahenu2 mice, restoration of liver PAH activity to only 10% of wild-type levels completely normalized serum phenylalanine levels in the animals [15]. Therefore, the amount of PAH activity in bone marrow of a double transgenic Tg/Pahenu2 mouse (approximately 15% of the total activity in a wild-type liver) should have been sufficient to clear phenylalanine from liver PAH-deficient mice, yet serum phenylalanine levels in these mice remained elevated. This result suggests that phenylalanine clearance in these mice was limited by some factor other than the absolute amount of PAH activity in marrow cells. However, this experiment does not rule out the possibility that much higher levels of PAH activity in marrow expressed from a more powerful promoter could effect a phenotypic change.
Although the amount of BH4 present natively in bone marrow should have been sufficient to support physiologically meaningful phenylalanine hydroxylation in Tg/Pahenu2 mice, we evaluated the hypothesis that phenylalanine clearance was still limited by BH4 supply. Following administration of BH4 by intraperitoneal injection, the BH4 content in marrow cells did increase, but serum phenylalanine levels of Tg/Pahenu2 mice were not altered. This suggests that BH4 supply was not limiting phenylalanine clearance in Tg/Pahenu2 mice.
Insufficient total flux of phenylalanine-containing plasma through the PAH-expressing marrow compartment is another possible cause of poor phenylalanine clearance in Tg/Pahenu2 mice. Little information concerning the rate of blood flow through the marrow compartment is available in the scientific or medical literature. Transferrin-bound 59Fe injected intravenously is predominantly cleared by erythroblasts in the bone marrow with some uptake occurring into reticuloendothelial cells of the liver and spleen [16]. The plasma 59Fe clearance rate provides an estimate of the rate of plasma flux through the marrow compartment. The plasma half-life of the isotope of a healthy adult varies from 60–140 min [17]. This compares to a plasma half-life of 12 min for intravenously injected BH4. These data suggest a rather slow flow rate of plasma through the marrow compartment although this interpretation could be complicated by possible limitations imposed by the kinetics of transferrin uptake into marrow cells. In our experiment, marrow uptake of intravenously administered BH4 was delayed, requiring 30 min before reaching its maximum. In comparison, BH4 uptake into skeletal muscle peaked at 5 min following injection. Interpretation of these data as a measure of blood flow into the marrow compartment is complicated by the fact that the majority of administered BH4 is cleared very rapidly from plasma by the liver and kidney. Still, we cannot rule out the possibility that phenylalanine clearance in Tg/Pahenu2 mice was limited by insufficient flux of phenylalanine-containing plasma through the PAH-expressing marrow compartment.
The rate of phenylalanine hydroxylation in intact marrow cells was significantly limited in comparison with marrow lysates. In contrast, the total capacity of liver to produce tyrosine did not substantially differ whether measured in liver homogenate or in intact isolated hepatocytes. This result suggests that hepatocytes and PAH-expressing LCRPAH marrow cells are fundamentally different in their ability to metabolize phenylalanine. Phenylalanine uptake across erythrocyte membranes has been studied extensively [18] and is probably not responsible for this difference. Phenylalanine transport across cell membranes is mediated by the sodium-independent system L transporter, which also mediates cellular uptake of other neutral amino acids. The system L transporter in the human erythrocyte has a very high affinity for (Km = 4.3 mM) and capacity for rapid transport (Vmax = 1.5 mmol/l cell water/min) of phenylalanine [19]. Although no specific data exist detailing phenylalanine transport into marrow erythroid precursors, there is no reason to suspect that the developing erythrocyte should not have an amino acid uptake system at least as robust as that of the mature red blood cell. Extracellular phenylalanine, whether in the in vitro PAH assay or in marrow of Tg/Pahenu2 mice, should have had free access to PAH enzyme contained within erythroid cells.
Cellular pterin uptake occurs via passive diffusion only [20], so insufficient intracellular supply of synthetic 6-methyltetrahydropterin cofactor could have limited our ability to measure phenylalanine hydroxylation in intact marrow cells. However, this limitation was not apparent in PAH assay of isolated hepatocytes, and increasing the cofactor concentration 10-fold in the PAH assay for marrow cells did not improve tyrosine production. Likewise, as discussed above, the intracellular concentration of BH4 in marrow should have been sufficient to support phenylalanine hydroxylation in Tg/Pahenu2 mice. The cause of the difference in tyrosine production rates between intact hepatocytes and LCRPAH marrow cells remains unknown. We cannot rule out the possibility that higher levels of PAH expression in marrow could overcome these limitations and yield physiologically significant phenylalanine clearance rates.
The results of our study, if confirmed, have profound implications for bone-marrow-directed heterologous gene therapy as a potential treatment for PKU or any other IEM. The efficacy of most contemporary gene therapy protocols is primarily limited by difficulties in producing and maintaining long-term, physiologically significant gene expression. Even if this problem is successfully surmounted, other factors may limit phenylalanine clearance from PAH-expressing bone marrow. Export into the circulation of mature hematopoietic cells exhibiting stable enzyme expression could potentially overcome this limitation; maintaining enzyme activity in circulating erythrocytes will, however, require the development of means to avoid the proteolytic processing inherent in erythroblast maturation. Further investigations, to include boosting PAH expression levels in erythrocytic marrow or by targeting myelopoietic cells, will be necessary to conclusively determine whether bone marrow could be an effective platform for the metabolism of phenylalanine in phenylketonuria.
Acknowledgements
CO Harding was supported by NIDDK K08 DK02405 and RO1 HD59371. The authors wish to thank Dr. Sheldon Milstien, Bethesda, MD, USA, for measurement of tissue BH4 levels and for insightful discussion concerning phenylalanine and BH4 metabolism.
References
- 1.Scriver CR, Kaufman S. Hyperphenylalaninemia: phenylalanine hydroxylase deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2001. pp. 1667–1724. [Google Scholar]
- 2.McDonald JD, Bode VC, Dove WF, Shedlovsky A. Pahhph-5: a mouse mutant deficient in phenylalanine hydroxylase. Proc Natl Acad Sci U S A. 1990;87:1965–1967. doi: 10.1073/pnas.87.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrester WC, Novak U, Gelinas R, Groudine M. Molecular analysis of the human beta-globin locus activation region. Proc Natl Acad Sci U S A. 1989;86:5439–5443. doi: 10.1073/pnas.86.14.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wet JR, Wood KV, DeLuca M, Helinski DR, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ledley FD, Grenett HE, Dunbar BS, Woo SL. Mouse phenylalanine hydroxylase. Homology and divergence from human phenylalanine hydroxylase. Biochem J. 1990;267:399–405. doi: 10.1042/bj2670399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harding CO, Wild K, Chang D, Messing A, Wolff JA. Metabolic engineering as therapy for inborn errors of metabolism – development of mice with phenylalanine hydroxylase expression in muscle. Gene Ther. 1998;5:677–683. doi: 10.1038/sj.gt.3300653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCaman MW, Robins E. Fluorimetric method for the determination of phenylalanine in serum. J Lab Clin Med. 1962;59:885–890. [Google Scholar]
- 8.Ledley FD, Hahn T, Woo SLC. Selection for phenylalanine hydroxylase activity in cells transformed with recombinant retrovirus. Somat Cell Mol Genet. 1987;13:145–154. doi: 10.1007/BF01534694. [DOI] [PubMed] [Google Scholar]
- 9.Overturf K, Al-Dhalimy M, Tanguay R, et al. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nature Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 10.Fukushima T, Nixon JC. Analysis of reduced forms of biopterin in biological tissues and fluids. Anal Biochem. 1980;102:176–188. doi: 10.1016/0003-2697(80)90336-x. [DOI] [PubMed] [Google Scholar]
- 11.Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brinster R, Chen H, Trumbauer M, Yagle M, Palmiter R. Factors affecting the efficiency of introducing foreign genes into mice by microinjecting eggs. Proc Natl Acad Sci U S A. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen R, Kolvraa S, Blaese RM, Jensen TG. Development of a skin-based metabolic sink for phenylalanine by overexpression of phenylalanine hydroxylase and GTP cyclohydrolase in primary human keratinocytes. Gene Ther. 2000;7:1971–1978. doi: 10.1038/sj.gt.3301337. [DOI] [PubMed] [Google Scholar]
- 14.Lin CM, Tan Y, Lee YM, Chang CC, Hsiao KJ. Expression of human phenylalanine hydroxylase activity in T lymphocytes of classical phenylketonuria children by retroviral-mediated gene transfer. J Inherit Metab Dis. 1997;20:742–754. doi: 10.1023/a:1005303331218. [DOI] [PubMed] [Google Scholar]
- 15.Fang B, Eisensmith RC, Li XHC, et al. Gene therapy for phenylketonuria: phenotypic correction in a genetically deficient mouse model by adenovirus-mediated hepatic gene therapy. Gene Ther. 1994;1:247–254. [PubMed] [Google Scholar]
- 16.Ricketts C, Jacobs A, Cavill I. Ferrokinetics and erythropoiesis in man: the measurement of effective erythropoiesis, ineffective erythropoiesis and red cell lifespan using 59Fe. Br J Haematol. 1975;31:65–75. doi: 10.1111/j.1365-2141.1975.tb00833.x. [DOI] [PubMed] [Google Scholar]
- 17.Peters A, Lewis S. Iron Metabolism. In: Maisey M, Britton K, Gilday D, editors. Clinical Nuclear Medicine. JB Lippincott Co.; Philadelphia: 1991. pp. 355–361. [Google Scholar]
- 18.Christensen HN. Interorgan amino acid nutrition. Physiol Rev. 1982;62:1193–1233. doi: 10.1152/physrev.1982.62.4.1193. [DOI] [PubMed] [Google Scholar]
- 19.Winter CG, Christensen HN. Migration of amino acids across the membrane of the human erythrocyte. J Biol Chem. 1964;239:872–878. [PubMed] [Google Scholar]
- 20.Anastasiadis PZ, Kuhn DM, Levine RA. Tetrahydrobiopterin uptake into rat brain synaptosomes, cultured PC12 cells, and rat striatum. Brain Res. 1994;665:77–84. doi: 10.1016/0006-8993(94)91154-1. [DOI] [PubMed] [Google Scholar]