Abstract
Myocardin belongs to the SAP domain family of transcription factors and is expressed specifically in cardiac and smooth muscle during embryogenesis and in adulthood. Myocardin functions as a transcriptional coactivator of SRF and is sufficient and necessary for smooth muscle gene expression. However, the in vivo function of myocardin during cardiogenesis is not completely understood. Here we clone myocardin from chick embryonic hearts and show that myocardin protein sequences are highly conserved cross species. Detailed studies of chick myocardin expression reveal that myocardin is expressed in cardiac and smooth muscle lineage during early embryogenesis, similar to that found in mouse. Interestingly, the expression of myocardin in the heart was found enriched in the outflow tract and the sinoatrial segments shortly after the formation of linear heart tube. Such expression pattern is also maintained in later developing embryos, suggesting that myocardin may play a unique role in the formation of those cardiac modules. Similar to its mouse counterpart, chick myocardin is able to activate cardiac and smooth muscle promoter reporter genes and induce smooth muscle gene expression in nonmuscle cells. Ectopic overexpression of myocardin enlarged the embryonic chick heart. Conversely, repression of the endogenous chick myocardin using antisense oligonucleotides or a dominant negative mutant form of myocardin inhibited cardiogenesis. Together, our data place myocardin as one of the earliest cardiac marker genes for cardiogenesis and support the idea that myocardin plays an essential role in cardiac gene expression and cardiogenesis.
Keywords: myocardin, SRF, heart, cardiac development, gene expression, transcription factor, chick embryos
The heart is the first organ to form during mammalian development (Srivastava and Olson, 2000; Olson, 2002). Several transcription factors have been implicated in activation of cardiac muscle gene expression during cardiomyocyte differentiation (Olson, 2004). NKX2.5, a homeobox protein, is expressed specifically in the heart and has been shown to bind a regulatory element in several cardiac gene control regions (Komuro and Izumo, 1993; Lints et al., 1993; Lyons et al., 1995; Tanaka et al., 1999). MEF2C, a member of myocyte enhance factor-2 (MEF2) family of MADS-box transcription factors, binds to a conserved A/T-rich DNA sequence in the control regions of the majority of cardiac, skeletal, and smooth muscle genes and is essential for cardiogenesis (Lin et al., 1997; Black and Olson, 1998; McKinsey et al., 2002). GATA4, a GATA family zinc finger protein which directly binds to the DNA sequence element (A/T)GATA(A/G), is expressed in the cardiac lineage throughout embryonic development and in adulthood, and is required for early heart formation (Arceci et al., 1993; Kelley et al., 1993; Ip et al., 1994; Kuo et al., 1997; Molkentin et al., 1997). Tbx1, Tbx5, and Tbx20, members of the T-box transcription factors, have also been shown as important regulators for cardiac gene expression and heart development (Horb and Thomsen, 1999; Bruneau et al., 2001; Hiroi et al., 2001; Merscher et al., 2001; Brown et al., 2003, 2005; Bussen et al., 2004; Hu et al., 2004; Singh et al., 2005; Stennard et al., 2005; Takeuchi et al., 2005). Mutations in some of those key transcription factors have been linked to human cardiovascular-related disease (Basson et al., 1997; Li et al., 1997; Epstein, 2001; Baldini, 2002).
We have recently identified a family of SAP domain transcription factors, myocardin and myocardin-related transcription factors (MRTFs) (Wang et al., 2001, 2002; Wang and Olson, 2004). Unlike other cardiac transcription factors, which bind to the conserved DNA sequences on the regulatory regions of their target gene, myocardin does not bind DNA alone, but forms a stable ternary complex with serum response factor (SRF) bound to the DNA consensus sequence CC(A/T)6GG, known as a CArG box. This interaction brings the powerful transcription activation domain (TAD) of myocardin to SRF-dependent target genes with resulting transcriptional activation (Wang and Olson, 2004; Wang et al., 2004b). Target genes that can be significantly transactivated by myocardin include cardiac-specific gene atrial natriuretic factor (ANF) and smooth muscle-specific gene SM22, both known targets for SRF (Pipes et al., 2006). In addition, myocardin is a key component of a molecular switch that regulates the ability of SRF to mediate cellular proliferation and muscle cell differentiation (Wang et al., 2004b; Liu et al., 2005).
As a transcriptional cofactor of SRF, myocardin potently transactivates CArG box-containing reporter genes (Wang et al., 2001). The functions of myocardin in smooth muscle gene expression and vascular smooth muscle cell development have been well documented (Chen et al., 2002; Du et al., 2003; Wang et al., 2003). Studies of myocardin knockout mice showed embryonic lethality without the formation of vascular smooth muscle cells (Li et al., 2003). However, the involvement of myocardin in cardiac development remains elusive. In Xenopus, whereas a dominant negative mutant of myocardin interfered cardiogenesis, overexpression of myocardin was able to ectopically induce cardiac gene expression (Wang et al., 2001; Small et al., 2005). Surprisingly, the heart in myocardin knockout mouse embryos seems to develop normally with an apparent normal expression of cardiac marker genes (Li et al., 2003).
To further clarify the potential role of myocardin in cardiogenesis, we cloned the avian ortholog of mammalian myocardin. Here we show that chick myocardin is specifically expressed in the cardiac lineage during early embryogenesis, similar to that found in mouse. Interestingly, we found that myocardin expression in the developing heart is not evenly distributed, instead, the highest myocardin expression is detected in the outflow tract and the sinoatrial segments. Like its mammalian counterpart, the chick myocardin is able to potently activate cardiac and smooth muscle gene expression as well as induce smooth muscle gene expression in nonmuscle cells. Whereas ectopic overexpression of myocardin seems to promote cardiac differentiation, repression of myocardin activity dramatically inhibits cardiogenesis in chick embryos. Our data establish myocardin as an early cardiac marker and support the view that myocardin plays an essential role in cardiogenesis.
MATERIALS AND METHODS
Cloning of Chick Myocardin
We used the mouse myocardin protein sequence to search the NCBI database (EST) and found one chick EST clone of 669 bp (GenBank access # BU143596) with high homology. Using this chick cDNA as a probe to screen a Hamburg-Hamilton (HH) Stage 21 embryonic chick heart cDNA library (a generous gift of Dr. Jim Lin, University of Iowa), we identified several overlapped cDNA clones from 500 bp to 1.5 kb. Sequencing of those cDNAs confirmed that they are partial sequences of the deduced chick myocardin gene. Using the most 5′-sequence of these cDNAs as a probe to rescreen, the cDNA library did not yield any additional upstream sequence information. To clone the full-length chick myocardin sequence, we used the mouse myocardin protein sequence to blast the chick genomic sequence database and found the putative chick myocardin genomic sequence on chromosome 18. This information allows us to deduce a chick myocardin full-length cDNA sequence. Based on the above information, we designed PCR primers which span over the putative chick myocardin open reading frame (ORF) sequence. Multiple cDNA fragments were PCR-amplified from cDNA pools derived from HH Stage 25 chick embryonic hearts and gizzard. Multiple cDNA fragments were amplified and subcloned into a pcDNA expression vector with a N-terminal Myc tag. All the cDNA clones were confirmed by sequencing.
Whole Mount In Situ Hybridization
Whole mount in situ hybridization was essentially performed as described previously (Wang et al., 1999) with minor modification. Two different cDNA fragments, which cover distinct portions of the chick myocardin transcripts (ORF and 3′ UTR), were used as templates for in vitro transcription to make antisense DIG-labeled riboprobes. These two probes were used to hybridize with staged chick embryos and yielded similar results. A sense-oriented riboprobe was used as a negative control. All experiments were repeated at least twice and multiple embryos were used for each developmental stage.
Cell Culture and Transfection Assays
Transfection of COS and 10T1/2 cells and luciferase reporter assays were performed as described (Wang et al., 2001, 2003, 2004b). Unless otherwise indicated, 100 ng of reporter plasmid and 100 ng of each activator plasmid were used. The total amount of DNA per well was kept constant by adding the corresponding amount of expression vector without a cDNA insert. All the transfection experiments were repeated at least twice in duplication. CMV-lacZ was included as an internal control for variations in transfection efficiency.
The mouse myocardin expression plasmids have been described (Wang et al., 2001, 2003, 2004b). Unless otherwise indicated, full-length chick and mouse myocardin expression plasmids were used. For chick myocardin expression constructs, varieties of chick myocardin cDNA fragments amplified from embryonic heart and gizzard cDNA pools were cloned into the pcDNA3.1 expression vector with a N-terminal Myc tag. To determine whether the constructs encode for myocardin proteins with correct size, the expression plasmids were transfected into the COS cells and the protein expression was confirmed by Western blot analysis. Myocardin deletion mutants were generated through PCR-based mutagenesis using the QuickChange kit (Stratagen). All mutations were confirmed by DNA sequencing. The SM22-luciferase reporter contained the 1,434 bp promoter and the ANF-luciferase reporter contained the 638 bp promoter (Wang et al., 2001).
Immunohistochemistry and Whole Mount Immunostaining of Chick Embryos
Myogenic conversion assays in 10T1/2 cells were performed as described (Wang et al., 2003) except that Lipofectamine reagent (Invitrogen) was used for transfection. After transfection, 10T1/2 cells were shifted to differentiation medium (DMEM plus 2% horse serum) for 48 hr before analysis. Mouse anti-SM-α-actin monoclonal antibody (1A4, Sigma) was used to monitor smooth muscle gene induction.
Whole mount immunostaining of chick embryos was performed primarily according to (Corson et al., 2003) with some modifications. After incubation with primary antibody (MF20, 1:5, Developmental Study Hybridoma Bank, Department of Biological Sciences, University of Iowa), an alkaline phosphatase (AP)-coupled secondary antibody was applied and color reaction was developed using the BM-Purple substrate (Roche).
Antisense Treatment and Microelectroporation
Antisense treatment and expression vector microelectroporation were performed as described previously (Wang et al., 2004a). Briefly, fertilized chick eggs (CBT Farm, Chestertown, MD) were incubated to approximately HH Stage 5–6. The embryos were explanted in new cultures and subjected to antisense treatment or microelectroporation. Antisense oligonucleotides were designed using IDT SciTools and synthesized as phosphorothioate derivatives and purified by HPLC (IDT, Corralville, IA). The sequence for the 20 bp antisense oligonucleotide that targets the C-terminus of the chick myocardin is 5′-ACTCCTCTCCTCCCATCTTC-3′. The inverse of the above sequence was used as control. Staged chick embryos were treated with 10 μL oligonucleotide mixed with lipofectamin (Invitrogen) at the concentration of 40 μM. For microelectroporation, a gold-plated cathode was fixed in the bottom of a 60-mm dish with a thin layer of Ringer’s saline, and Stage 6 embryos were prepared for new culture and put onto the cathode. About 1 μg of expression plasmid myocardin or dominant negative myocardin mixed with pMES, or pMES alone as control, was injected into the target site between the blastoderm and vitelline membrane using a glass capillary. A platinum anode (0.5 mm in diameter) was then placed on the hypoblast at the prospective heart-forming region. The parameters for electroporation were 5 pulses of 5 V for the duration of 25 msec with the intervals of 454 msec. The treated embryos were placed on the agar media in new cultures and incubated in 38°C to the desired stages for immunostaining for gene expression. The expression of expression plasmids after microelectroporation was monitored by GFP expression of pMES plasmid.
RESULTS
Cloning of Avian Myocardin
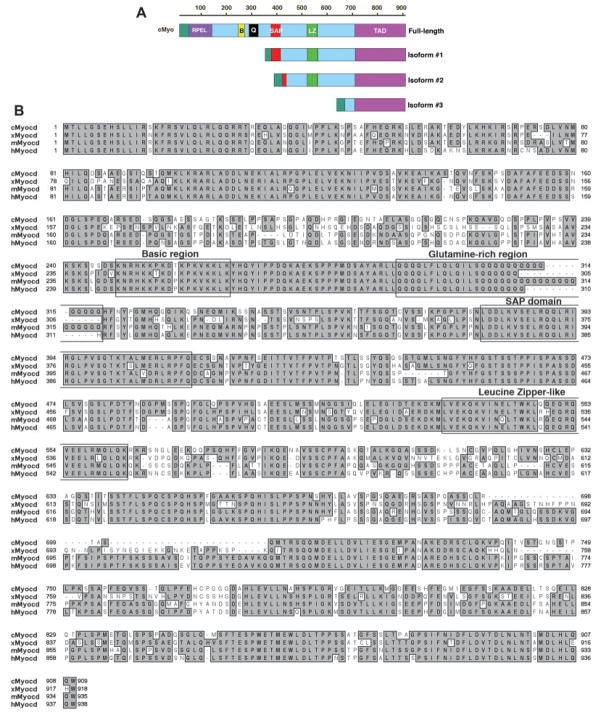
A partial chick myocardin cDNA sequence was identified when the mouse myocardin protein sequence was used to search the NCBI EST database. Using this chick myocardin cDNA as a probe to screen a chick embryonic heart cDNA library, we identified additional overlapped cDNA clones, representing chick myocardin sequences. Finally, a PCR-based cloning approach allowed us to clone the chick myocardin cDNA sequences (see Material and Methods). Among these cDNA clones, two appear to represent the full-length chick myocardin cDNA with a deduced peptide of 909 amino acids (Fig. 1).
Fig. 1.
Cloning of the chick myocardin and the comparison of myocardin protein sequences from chick, frog, mouse, and human. (A) Diagram of the chick myocardin isoforms cloned in this study. Protein structural domains are indicated as follows: RPEL, RPEL domain; B, basic domain; Q, glutamine-rich domain; SAP, SAP domain; LZ, leucine zipper-like domain; TAD, transcription activation domain. (B) Alignment of myocardin protein sequences from chick (cMyocd), frog (xMyocd), mouse (mMyocd), and human (hMyocd). Some of the important domains are labeled. The chick myocardin is 64%, 64%, and 67% identical to its frog, mouse, and human homologues, respectively.
In addition to the putative full-length chick myocardin cDNAs, we have also cloned several short isoforms of the chick myocardin gene, with the size ranged from 1.1 to 2.2 kb. The deduced amino acid sequences of these isoforms were aligned to the full-length protein (Fig. 1). Interestingly, most of those isoforms contain an intact C-terminal transactivation domain (TAD). Analysis against chick genomic sequences indicates that all of these short transcripts are the products of differential splicing from the same chick myocardin gene (data not shown). Using the chick myocardin protein sequence to blast the chick genomic database at decreased stringency, we found three hits on chick genomic loci on chromosomes 18, 14, and 1, respectively (data not shown). Given that we have previously identified three myocardin family proteins from mammals, myocardin, MRTF-A, and MRTF-B (Wang et al., 2002), those three chick genes are likely the avian homologues of myocardin family transcription factors.
Myocardin Protein Sequences Are Highly Conserved
Comparison of myocardin protein sequences cross species shows that the chick myocardin protein shares 64%, 64%, and 67% amino acid sequence identity to its frog, mouse, and human counterparts, respectively (Fig. 1B). The chick myocardin protein contains the predicted conserved domains found in other members of the myocardin family proteins. Those include the N-terminal RPEL domain (RPEL), the basic domain, glutamine-rich domain (Q-domain), SAP domain, leucine-zipper like domain (LZ), and the C-terminal transactivation domain (TAD) (Fig. 1). Aligning the chick myocardin cDNA sequence against the chick genomic sequences revealed that the chick myocardin has a very similar exon-intron structure as that of mouse myocardin gene (data not shown). Together, these data demonstrate that the myocardin proteins are highly conserved evolutionally.
Expression of Chick Myocardin During Embryogenesis
We and others have previously reported that myocardin is specifically expressed in cardiac and smooth muscle cells during mouse development and in adulthood (Wang et al., 2001; Chen et al., 2002; Du et al., 2003; Ueyama et al., 2003). Most of those previous experiments were performed using in situ hybridization on mouse embryo tissue sections. Unfortunately, such approach was not an ideal approach to view the three-dimensional spatiotemporal expression pattern of myocardin. We have recently shown, using whole mount in situ hybridization, that Xenopus myocardin is expressed in the heart of frog embryos (Small et al., 2005). However, this study was limited to only very early developmental stages and Xenopus myocardin expression was not studied in whole mount frog embryos at later developmental stages (Wang et al., 2001; Small et al., 2005). We therefore decided to apply whole mount in situ hybridization to examine the expression patterns of the chick myocardin during embryogenesis. Two independent antisense riboprobes, which are from two different region of the chick myocardin cDNA, were used in the whole mount in situ hybridization and both yielded the same experimental results. Sense oriented probes served as negative controls and no signal was detected in those embryos (data not shown). The chick myocardin transcripts are first clearly detected in the cardiac crescent, the precursor of earliest cardiac lineage, of HH Stage 8–9 chick embryos (Fig. 2A). Myocardin expression was not detected in any other region of the embryos at this stage (Fig. 2A). At HH Stage 10, myocardin is expressed in the newly formed linear heart tube with an apparently homogenous expression level in the whole heart tube (Fig. 2B). Myocardin continues to be expressed in the looping heart of HH Stage 11 embryos. Interestingly, stronger myocardin expression is detected at the posterior portion of the developing embryonic heart, the future atrium and the sinus venosus (Fig. 2C,D). By HH Stage 12–13, myocardin expression is still restricted to the developing heart, but a much higher expression level of myocardin transcript is detected in the outflow tract and future atria (Fig. 2E–H). Cardiac-specific expression of myocardin continues in HH Stage 15 chick embryos, with higher expression in the outflow tract and atria (Fig. 2I–L, arrows). Similar myocardin expression pattern is maintained in HH Stage 18 chick embryos (Fig. 2M–P). At HH Stage 21, myocardin expression is detected in the heart where a high level of expression in the ventricle is evident (Fig. 2Q–T). Of a particular note, myocardin is also expressed in the developing allantoic vesicle, a smooth muscle enriched structure (Fig. 2Q). The expression patterns of chick myocardin in developing chick embryos are specific: we were able to obtain identical expression patterns from multiple independent experiments using multiple embryos. Together, these data establish cardiac-specific expression of the chick myocardin during development.
Fig. 2.
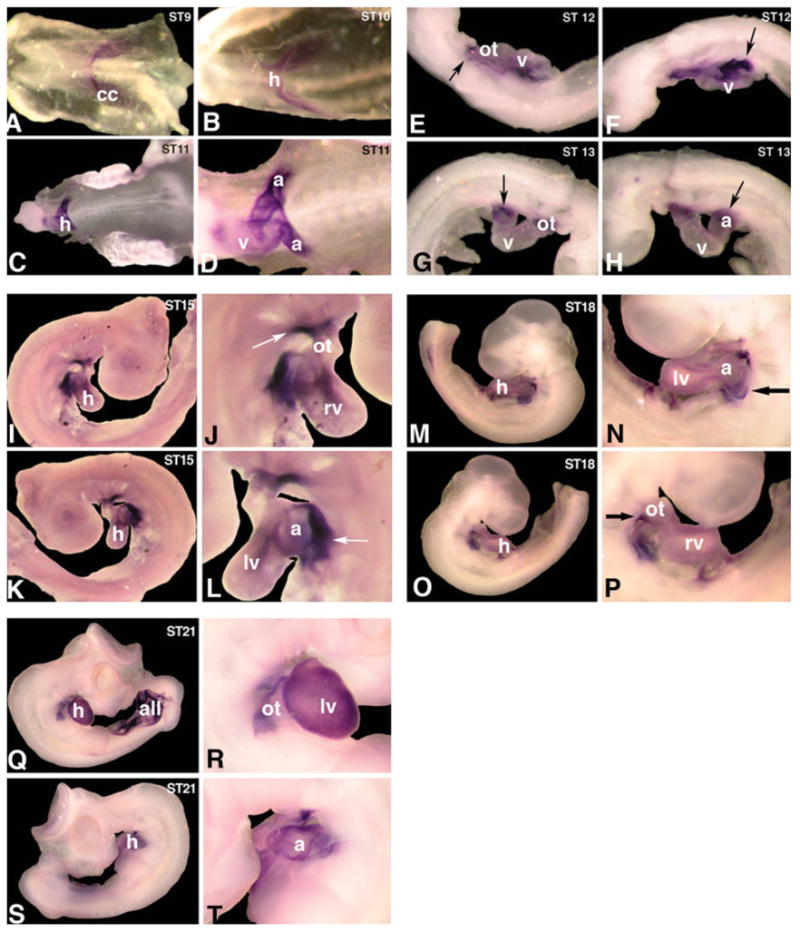
Expression of chick myocardin transcripts in chick embryos. Whole mount in situ hybridization was used to detect the expression of chick myocardin transcripts in staged chick embryos during early developmental stages. Myocardin expression was first detected in the cardiac crescent (cc) of Hamburger-Hamilton (HH) Stage 9 chick embryos (A). Myocardin is expressed in the newly formed heart tube (h) of HH Stage 10 and 11 embryos (B and C). Higher level of myocardin expression was detected in the future atrium (a) of HH Stage 11 embryo (D). Myocardin expression is restricted to the cardiac lineage in HH Stage 12–13 chick embryos (E–H). Arrows indicate higher level of myocardin expression detected in the atrium (a) and outflow tract (ot). Myocardin expression in the hearts of HH Stage 15 chick embryos (I–L). Arrows indicate higher level of myocardin expression in the atrium (a) and outflow tract (ot). Myocardin expression in HH Stage 18 chick embryos (M–P). Arrows indicate higher level of myocardin expression detected in the atrium (a) and outflow tract (ot). Myocardin is expressed in the developing heart (h) and allantoic vesicle (all) of HH Stage 21 chick embryos (Q–T). Images in 2D, J, L, N, P, R, and T represent higher magnification of 2C, I, K, M, O, Q, and S, respectively. Abbreviations: a, atrium; all, allantoic vesicle; cc, cardiac crescent; h, heart; lv, left ventricle; ot, outflow tract; rv, right ventricle; v, ventricle.
To compare the observed chick myocardin expression patterns with that of its mouse counterpart, we performed whole mount in situ hybridizations using a mouse myocardin riboprobe on embryonic day 9.5 mouse embryos (E9.5). The mouse myocardin expression pattern is very similar to that of the chick myocardin (Fig. 3). The highest expression was detected in the outflow tract and atrium. Intriguingly, we have consistently observed that the expression of the mouse myocardin in a restricted region of the forebrain of the staged mouse embryos (Fig. 3B). These results are particularly interesting, given a recent report showing that SRF and myocardin are involved in smooth muscle gene expression and cerebral blood flow of Alzhemer’s patients (Chow et al., 2007).
Fig. 3.
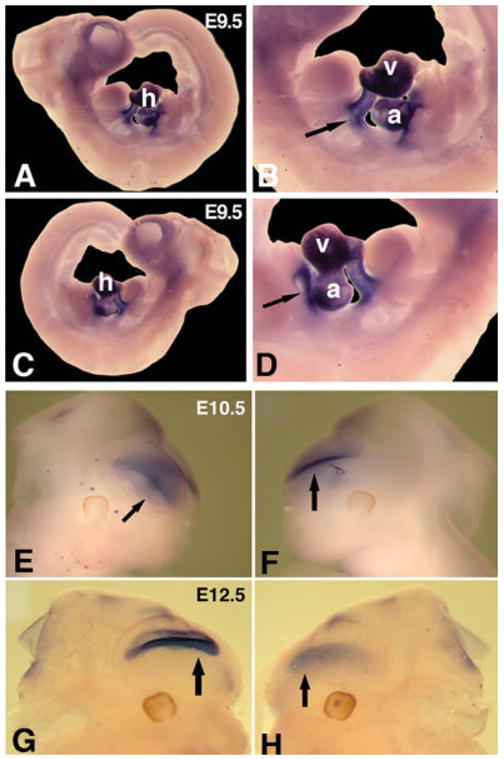
Expression of mouse myocardin transcripts in mouse embryos. Whole mount in situ hybridization was used to detect the expression of mouse myocardin transcripts in staged mouse embryos. A–D: Mouse myocardin is expressed in the hearts of embryonic day 9.5 (E9.5) mouse embryos with higher expression in the outflow tract and atrium (arrows). E–H: Mouse myocardin expression in a restricted region of anterior asects of E10.5 (E and F) and E12.5 (G and H) mouse embryos. Abbreviations: a, atrium; h, heart; v, ventricle.
Activation of Cardiac and Smooth Muscle Reporter Genes by Chick Myocardin
We have previously demonstrated that the mouse myocardin was able to potently activate cardiac and smooth muscle target gene expression (Wang et al., 2001). The high conservation between the mouse and chick myocardin protein sequences as well as the similarity in their expression patterns during embryogenesis suggest that the chick myocardin may function in a similar fashion. We therefore tested if the chick myocardin is able to activate cardiac reporter gene ANF and smooth muscle reporter gene SM22, using transfection luciferase reporter assays in cultured mammalian cells. Expression plasmids for full-length chick myocardin potently activate both the ANF and SM22 luciferase reporters to a level comparable to that of the mouse myocardin (Fig. 4A,B). However, the chick myocardin short isoforms were not able to activate those reporter genes (Fig. 4A,B). Since most of these chick myocardin short isoforms lack the N-terminal region of myocardin protein, which is highly evolutionary conserved, these data suggest that the N-terminal portion of the myocardin protein is essential for its function. Those data are consistent with our previous reports in which we showed that the specificity of the mouse myocardin protein is determined by its N-terminus (Wang et al., 2001, 2004b; Wang and Olson, 2004; Cao et al., 2005).
Fig. 4.
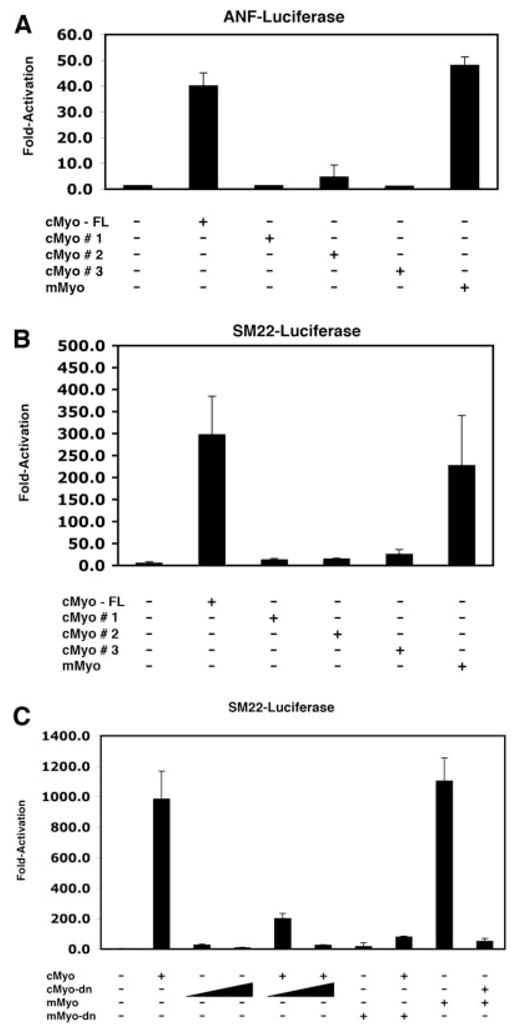
Activation of cardiac and smooth muscle reporter genes by myocardin proteins. COS cells were transfected with ANF or SM22 luciferase reporters and the indicated chick or mouse myocardin expression constructs. Luciferase activities were measured and results were presented as fold-activation from three independent experiments in duplication. (A) Activation of the ANF promoter luciferase reporter by chick myocardin (cMyo). Only the full-length cMyo, but not the short isoforms, was able to activate ANF reporter. Mouse myocardin (mMyo) was used for comparison is this assay. (B) Activation of the SM22 promoter luciferase reporter by the full-length cMyo, not the short isoforms. (C) Dominant negative mutants of chick or mouse myocardin repress wild-type (full-length) myocardin-mediated transactivation of the SM22 promoter luciferase reporter.
We then examined whether a dominant negative mutant of mouse myocardin, which lacks its C-terminal TAD domain, was able to interfere with the transcriptional activity of the full-length chick myocardin. The mouse myocardin dominant negative mutant dramatically repressed the transcriptional activity of the chick myocardin, similar to its ability to repress the transcriptional activity of the full-length mouse myocardin (Fig. 4C; and Wang, et al., 2001). Similarly, a chick myocardin dominant negative mutant which lacks its TAD domain, potently suppressed the transactivities of both mouse and chick myocardin proteins (Fig. 4C). These results demonstrated that the chick myocardin is a potent transcription factor for cardiac and smooth muscle genes.
Activation of Smooth Muscle Genes by Chick Myocardin
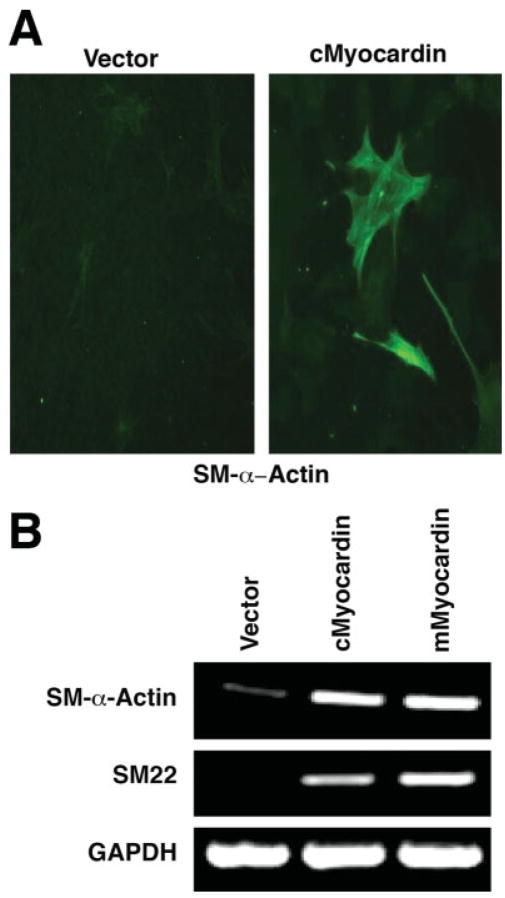
Next, we tested whether ectopic expression of the chick myocardin in nonmuscle 10T1/2 cells could activate smooth muscle gene expression. Similar to what we have observed previously for the mouse myocardin, smooth muscle gene expression is readily detected by immunostaining using a monoclonal antibody against smooth muscle α-actin after chick myocardin was transfected into the 10T1/2 cells (Fig. 5A). Semiquantitative RT-PCR analyses demonstrate that the chick myocardin is able to induce smooth muscle gene expression to a level comparable to that of the mouse myocardin when ectopically overexpressed in the 10T1/2 cells (Fig. 5B). Together, these data demonstrate that not only the protein structure and expression pattern but also the function of myocardin proteins are highly conserved.
Fig. 5.
Induction of smooth muscle gene expression by chick myocardin. A: 10T1/2 cells were transfected with chick myocardin (cMyocardin) expression construct. Twenty-four hours after transfection, the cells were shifted to differentiation medium (DMEM plus 2% horse serum) for 48 hr and smooth muscle α-actin (sm-α-actin) expression was assayed by immunostaining. Cells transfected with a pcDNA vector were used as a control (vector). B: 10T1/2 cells were transfected with chick myocardin (cMyocardin) or mouse myocardin (mMyocardin) expression construct. Twenty-four hours after transfection, the cells were shifted to differentiation medium (DMEM plus 2% horse serum) for 48 hr. Total RNAs were isolated and the expression of smooth muscle genes (SM-α-actin and SM22) were assayed using semiquantitative RT-PCR. GAPDH was measured as a loading control.
Myocardin Is Required for Cardiogenesis in Chick Embryos
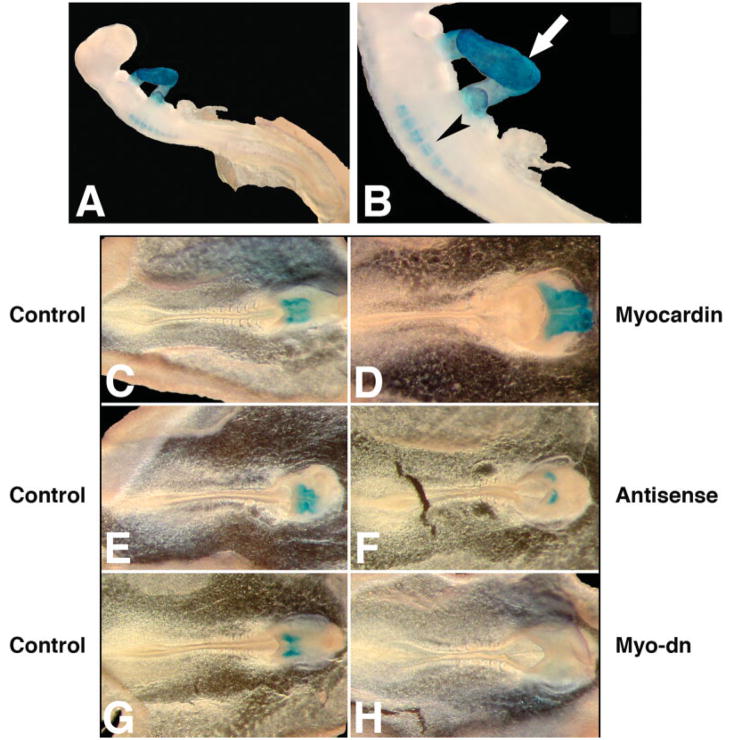
To investigate the in vivo function of myocardin during cardiac development, we set out to overexpress or knock down myocardin in cultured chick embryos. An expression plasmid for either chick or mouse myocardin was introduced into HH Stage 5–6 chick embryos using microelectroporation procedure (Wang et al., 2004a). An expression plasmid for GFP was coelectroporated in order to monitor the uptake and expression of introduced DNA. Processed embryos were cultured for additional 24 hr to reach HH Stage 10–11 before harvesting. Control and treated embryos were then proceeded for whole mount immunohistology analysis using a MF20 antibody, which specifically recognize cardiac and skeletal muscle myosin heavy chain (Fig. 6A,B; and Chen et al., 2006). Overexpression of myocardin in chick embryos caused an enlargement of embryonic hearts (61%, n = 18) (Fig. 6D). In contrast, control embryos with only a GFP expression plasmid did not show any change in their heart development (n = 15) (Fig. 6C). We did not observe ectopic MF20-positive tissues in any other region of the chick embryos other than the heart region. We did not obtain ectopic smooth muscle marker gene expression in those embryos (data not shown).
Fig. 6.
Myocardin is essential for cardiogenesis in chick embryos. A,B: Whole mount immunostaining of HH Stage 12 chick embryos using the MF20 monoclonal antibody, which specifically recognizes striated cardiac and skeletal muscle myosin heavy chain. The developing heart (arrow) and somites (arrowhead) are indicated. B: Higher magnification of (A). Chick embryos in new culture were microelectroporated with expression plasmids for either a wild-type myocardin (D) or a dominant negative mutant form of myocardin (H), or treated with antisense oligonucleotides specific for myocardin (F) or their respective controls (C, G, and E) as described in Materials and Methods. After incubation for additional 24 hr, embryos were harvested and analyzed by whole mount immunostaining using the MF-20 monoclonal antibody, which marks the cardiac structure (blue).
Conversely, introduction of a dominant negative mutant of myocardin into the chick embryos leads to a defect in cardiogenesis which is associated with the decrease or completed blockage of cardiac marker gene MHC expression (Fig. 6G,H and data not shown). Such observed heart development defects are specific because other aspects of embryonic development appear to be normal in those embryos. To further confirm the above observation, we applied an antisense oligonucleotide specific to the chick myocardin sequence to staged chick embryos in culture. Antimyocardin antisense oligonucleotide dramatically inhibits cardiac development and represses the expression of myosin heavy chain protein, as marked by MF-20 antibody staining. As a negative control, no inhibitory effect was observed in cultured chick embryos treated with a control oligonucleotide (Fig. 6E,F). Collectively, our data demonstrated that myocardin plays an essential role in cardiogenesis in chick embryos.
DISCUSSION
In this study, we have cloned the avian ortholog of the mammalian myocardin gene. We show that the chick myocardin is specifically expressed in the developing hearts of early chick embryos. Interestingly, we found that the expression of the chick myocardin in the heart is enriched in the outflow tract and atrium during early embryogenesis, indicating that myocardin may play a role in the morphogenesis of those modules. We have also demonstrated that chick myocardin, like its mammalian counterparts, is able to potently activate cardiac and smooth muscle gene expression. Most importantly, gain- and loss-of-function studies in chick embryos clearly established an important role of the myocardin protein in cardiac gene expression and heart development.
Expression of Myocardin During Heart Development
The transcripts of the chick myocardin are first detected in the cardiac crescent of chick embryos, making myocardin one of the earliest markers for cardiogenic specification. Our expression studies reveal that the expression of the chick myocardin is restricted to the cardiac lineage during early embryogenesis, consistent with previous studies in other species, including frog and mouse (Wang et al., 2001; Small et al., 2005). Unexpectedly, we found that the expression of myocardin is not homogenously distributed in the whole heart. Instead, a much higher level of myocardin expression was detected in the outflow tract and developing atria of chick embryos. Such myocardin expression pattern suggests that myocardin may play a specific role in the morphogenesis of outflow tract and atria.
Recently, the regulatory regions of the mouse myocardin gene which direct its cardiac- and smooth muscle-specific expression during embryogenesis and in adult animals were identified (Creemers et al., 2006a). It has been shown that the cardiac- and smooth muscle-specific expression of the mouse myocardin gene is controlled by the combination of MEF2, Foxo, and Tead transcription factors to bind to the myocardin enhancer. In particular, the MEF2 and Foxo proteins are responsive to the expression of myocardin in both cardiac and smooth muscle lineage, whereas the Tead transcription factors appear to be required for the expression of myocardin in the smooth muscle cells (Creemers et al., 2006a). It will be important to determine whether myocardin contributes to the developmental defects associated with the loss-of-function of Mef2, Foxo, or Tead proteins.
It is now recognized that two distinct sources of cardiac precursor cells, which come from the so-called “primary” and “secondary heart fields,” are needed for the cardiogenesis (Kirby, 2002; Buckingham et al., 2005). Whereas the primary heart field is essential for the formation of primary heart tube, additional cardiac precursor cells are recruited from the secondary heart field to contribute to the future right ventricle and outflow tract (Kelly et al., 2001; Cai et al., 2003). Loss-of-function studies in genes which are thought to play an important role in the secondary heart field fate clearly demonstrated that the secondary heart field is required for the morphogenesis of outflow tract and right ventricle (Srivastava et al., 1995, 1997; Lin et al., 1997; Kelly et al., 2001; Waldo et al., 2001; Cai et al., 2003; von Both et al., 2004). In this study, we found that the chick myocardin is expressed in the cardiac crescent of Stage 8 chick embryos. Later on, its expression in the developing heart is enriched in the outflow tract and atria. It will be extremely interesting to determine whether myocardin is also expressed in the secondary heat field and if it plays a role in the morphogenesis of right ventricle and outflow tract.
Function of Myocardin in Heart Development and Cardiac Gene Expression
Previously, we showed that myocardin plays a critical role during normal heart development, when a dominant-negative myocardin mutant was introduced into Xenopus embryos to inhibit the function of endogenous myocardin (Wang et al., 2001). Such conclusion was further supported by a recent study where myocardin was knocked down by morpholino oligos in Xenopus. Significantly, ectopic overexpression of myocardin in Xenopus was sufficient to induce ectopic cardiac gene expression, suggesting that myocardin may function as a key transcription factor to activate cardiac gene expression program (Small et al., 2005). Surprisingly, the early cardiogenesis appears to be normal in the myocardin knockout mouse embryos. This has been explained by the possible functional redundancy between the myocardin family of transcription factors (MRTF-A and MRTF-B) (Li et al., 2003). In this study, when the chick myocardin was introduced into early chick embryos, we observed an increase in heart size. However, we never observed ectopic cardiac marker gene induction outside the cardiac developmental region. It will be interesting to understand why evolution has specified the myocardin’s role in controlling cardiac development in such a way that a much more profound role is found in amphibians than in avians and mammals.
Myocardin Family of Transcription Factors and Their Function
In addition to myocardin, we have identified two MRTFs-A and -B from mouse, which share homology with the SAP domain, as well as the basic and glutamine-rich domains of myocardin (Wang et al., 2002). Myocardin and MRTFs comprise a novel family of SRF cofactors with extraordinary transcriptional potency (Wang and Olson, 2004). Whereas myocardin is expressed in a cardiac- and smooth muscle-specific manner, MRTF-A and MRTF-B are widely expressed in embryonic and adult tissues (Wang et al., 2002). Their different expression patterns and differential effects on SRF activity suggest that myocardin and MRTFs participate in distinct SRF-dependent programs of gene expression. Indeed, gene targeting studies reveal that MRTF-A is required for the differentiation of mammary myoepithelial cells (Li et al., 2005, 2006; Oh et al., 2005; Sun et al., 2006; Wei et al., 2007) and MRTF-B is essential for the correct patterning of branchial arch arteries and outflow tract (Li et al., 2005; Oh et al., 2005; Wei et al., 2007). Using the chick myocardin protein sequence to search the chick genome at decreased stringency, we found three genomic loci on chromosomes 18, 14, and 1, respectively, suggesting the presence of three myocardin family members in chick (data not shown). Surprisingly, when we used the chick or mouse myocardin protein sequences to search the genomes of fugu fish and zebra-fish, we identified four genomic loci in those organisms, suggesting the existence of one additional myocardin family member (Wang et al., unpublished observation).
Intriguingly, the mouse myocardin was found to interact with MEF2 proteins via a unique N-terminal peptide sequence. We have found that such MEF2 interacting domain is highly conserved in the chick myocardin. Furthermore, such unique MEF2-binding domain of mouse myocardin was used to search for additional MEF2 interacting proteins and has lead to the identification of MASTR, a novel SAP domain protein and coactivator of MEF2 proteins. MASTR was shown to synergize with MEF2 to activate myogenic gene expression in skeletal muscle (Creemers et al., 2006b). However, MASTR is unlikely a new member of myocardin family transcription factor, even thought it is a member of SAP domain containing nuclear proteins. MASTR does not contain the signature domains found in other myocardin family members (Basic, Q-domain, Leucine Zipper). Clearly, the discovery of myocardin family of transcription factors and related proteins has provided opportunity to investigate the molecular mechanisms of cell growth and differentiation. With the cloning and characterization of the chick myocardin, it will be important in the future to study the upstream signals and downstream regulatory target genes which mediate the function of myocardin in muscle differentiation and muscle gene expression.
Acknowledgments
National Institutes of Health, March of Dimes Birth Defects Foundation, Muscular Dystrophy Association, American Heart Association.
We thank Jim Lin (University of Iowa) for the chick embryonic heart cDNA library. Mark Majesky for allowing us to use his egg incubators and microscopes. Noah Park for participation in this work at the initial stage and James Ouyang for graphics. We thank Mark Mejesky, Frank Conlon, and Tom Callis for their careful reading of this manuscript. We also appreciate members of the Wang laboratory for discussion. MF20 antibody was obtained from the developmental studies hybridoma bank (Department of Biological Sciences, University of Iowa, Iowa City, IA). Drs. Y Chen and D.Z Wang are established investigators of the American Heart Association.
LITERATURE CITED
- Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993;13:2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini A. DiGeorge syndrome: the use of model organisms to dissect complex genetics. Hum Mol Genet. 2002;11:2363–2369. doi: 10.1093/hmg/11.20.2363. [DOI] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Brown DD, Binder O, Pagratis M, Parr BA, Conlon FL. Developmental expression of the Xenopus laevis Tbx20 orthologue. Dev Genes Evol. 2003;212:604–607. doi: 10.1007/s00427-002-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Martz SN, Binder O, Goetz SC, Price BM, Smith JC, Conlon FL. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132:553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Bussen M, Petry M, Schuster-Gossler K, Leitges M, Gossler A, Kispert A. The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes Dev. 2004;18:1209–1221. doi: 10.1101/gad.300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Wang Z, Zhang CL, Oh J, Xing W, Li S, Richardson JA, Wang DZ, Olson EN. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Mol Cell Biol. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow N, Bell RD, Deane R, Streb JW, Chen J, Brooks A, Van Nostrand W, Miano JM, Zlokovic BV. Serum response factor and myocardin mediate arterial hypercontractility and cerebral blood flow dysregulation in Alzheimer’s phenotype. Proc Natl Acad Sci USA. 2007;104:823–828. doi: 10.1073/pnas.0608251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN. Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development. 2006a;133:4245–4256. doi: 10.1242/dev.02610. [DOI] [PubMed] [Google Scholar]
- Creemers EE, Sutherland LB, Oh J, Barbosa AC, Olson EN. Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol Cell. 2006b;23:83–96. doi: 10.1016/j.molcel.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA. Developing models of DiGeorge syndrome. Trends Genet. 2001;17:S13–S17. doi: 10.1016/s0168-9525(01)02450-7. [DOI] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. Tbx5 is essential for heart development. Development. 1999;126:1739–1751. doi: 10.1242/dev.126.8.1739. [DOI] [PubMed] [Google Scholar]
- Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D. Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development. 2004;131:5491–5502. doi: 10.1242/dev.01399. [DOI] [PubMed] [Google Scholar]
- Ip HS, Wilson DB, Heikinheimo M, Tang Z, Ting CN, Simon MC, Leiden JM, Parmacek MS. The GATA-4 transcription factor transactivates the cardiac muscle-specific troponin C promoter-enhancer in nonmuscle cells. Mol Cell Biol. 1994;14:7517–7526. doi: 10.1128/mcb.14.11.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley C, Blumberg H, Zon LI, Evans T. GATA-4 is a novel transcription factor expressed in endocardium of the developing heart. Development. 1993;118:817–827. doi: 10.1242/dev.118.3.817. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- Kirby ML. Molecular embryogenesis of the heart. Pediatr Dev Pathol. 2002;5:516–543. doi: 10.1007/s10024-002-0004-2. [DOI] [PubMed] [Google Scholar]
- Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, Du K, Epstein JA, Parmacek MS. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci USA. 2005;102:8916–8921. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, Bonnet D, Lyonnet S, Young ID, Raeburn JA, Buckler AJ, Law DJ, Brook JD. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- Li S, Chang S, Qi X, Richardson JA, Olson EN. Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol Cell Biol. 2006;26:5797–5808. doi: 10.1128/MCB.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci USA. 2003;100:9366–9370. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:969. doi: 10.1242/dev.119.3.969. [DOI] [PubMed] [Google Scholar]
- Liu ZP, Wang Z, Yanagisawa H, Olson EN. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev Cell. 2005;9:261–270. doi: 10.1016/j.devcel.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Oh J, Richardson JA, Olson EN. Requirement of myocardin-related transcription factor-B for remodeling of branchial arch arteries and smooth muscle differentiation. Proc Natl Acad Sci USA. 2005;102:15122–15127. doi: 10.1073/pnas.0507346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. A genetic blueprint for growth and development of the heart. Harvey Lect. 2002;98:41–64. [PubMed] [Google Scholar]
- Olson EN. A decade of discoveries in cardiac biology. Nat Med. 2004;10:467–474. doi: 10.1038/nm0504-467. [DOI] [PubMed] [Google Scholar]
- Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- Small EM, Warkman AS, Wang DZ, Sutherland LB, Olson EN, Krieg PA. Myocardin is sufficient and necessary for cardiac gene expression in Xenopus. Development. 2005;132:987–997. doi: 10.1242/dev.01684. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, Elliott DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- Sun Y, Boyd K, Xu W, Ma J, Jackson CW, Fu A, Shillingford JM, Robinson GW, Hennighausen L, Hitzler JK, Ma Z, Morris SW. Acute myeloid leukemia-associated Mkl1 (Mrtf-a) is a key regulator of mammary gland function. Mol Cell Biol. 2006;26:5809–5826. doi: 10.1128/MCB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, Hui CC, Henkelman RM, Nemer M, Black BL, Nagy A, Bruneau BG. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Ueyama T, Kasahara H, Ishiwata T, Nie Q, Izumo S. Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis. Mol Cell Biol. 2003;23:9222–9232. doi: 10.1128/MCB.23.24.9222-9232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL. Foxh1 is essential for development of the anterior heart field. Dev Cell. 2004;7:331–345. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A, Olson EN. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc Natl Acad Sci USA. 2002;99:14855–14860. doi: 10.1073/pnas.222561499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DZ, Olson EN. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr Opin Genet Dev. 2004;14:558–566. doi: 10.1016/j.gde.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Reiter RS, Lin JL, Wang Q, Williams HS, Krob SL, Schultheiss TM, Evans S, Lin JJ. Requirement of a novel gene, Xin, in cardiac morphogenesis. Development. 1999;126:1281–1294. doi: 10.1242/dev.126.6.1281. [DOI] [PubMed] [Google Scholar]
- Wang S, Yu X, Zhang T, Zhang X, Zhang Z, Chen Y. Chick Pcl2 regulates the left-right asymmetry by repressing Shh expression in Hensen’s node. Development. 2004a;131:4381–4391. doi: 10.1242/dev.01269. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004b;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Che N, Chen F. Myocardin-related transcription factor B is required for normal mouse vascular development and smooth muscle gene expression. Dev Dyn. 2007;236:416–425. doi: 10.1002/dvdy.21041. [DOI] [PubMed] [Google Scholar]