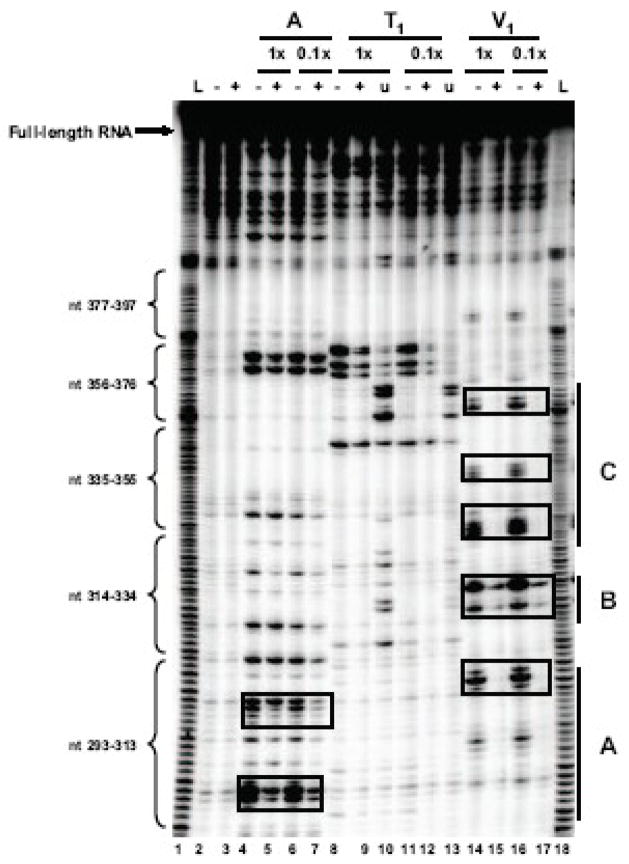
Figure 3. RNase foot-printing assay of coxsackievirus stem-loop IV RNA and PCBP2.

A.PCBP2 protection of multiple regions of coxsackievirus stem-loop IV RNA. Radio-labeled coxsackievirus stem-loop IV RNA (20–40 ng) was incubated with 5 μM of PCBP2, denoted by +, at 30°C for 10 minutes and subjected to RNase A (lanes 4–7), T1 (lanes 8–13) and V1 (lanes 14–17) digestion. The RNAs were digested for 1–3 minutes at 25°C with RNase dilution of 1:1 and 1:10 from stock. Urea (1 mM) was added to the T1 digestion to obtain a G-ladder (lanes 10 and 13). Boxes indicate nucleotides within coxsackievirus stem-loop IV RNA that were protected by PCBP2 from RNase cleavage. B. Secondary structure showing regions of sequence in coxsackievirus stem-loop IV RNA protected by PCBP2 from RNase digestion (denoted by boxes). PCBP2 protects the poly(C)-loop, the stem leading to the internal bulge, and the helix region adjacent to the poly(Py)-bulge.