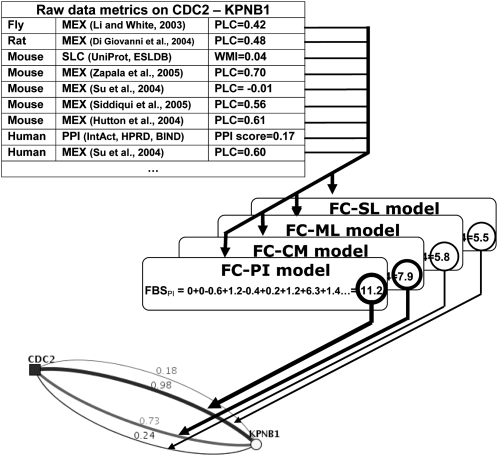
Figure 2.
Example of predicting functional coupling (FC) by FunCoup's naïve Bayesian network. To evaluate the cumulative likelihood of the link CDC2–KPNB1, evidence (only nine shown) from a number of species were available. The metrics computed from each evidence were scored by the four models of FC and summed up to a total likelihood value, the final Bayesian score (FBS). In this example, a physical protein interaction (FC-PI) is more likely than membership in the same protein complex (FC-CM), a metabolic (FC-ML), or a signaling (FC-SL) link. The web interface to the database of predictions (http://FunCoup.sbc.su.se) displays likelihoods of the FC classes as four colored lines with FBS values transformed into the pfc confidence scores. This coupling was supported by many evidences from several species (top). The strongest came from the IntAct database that reported two experiments (Thelemann et al. 2005; Koch et al. 2007) where CDC2 and KPNB1 were mentioned as preys interacting with the same bait (in parallel with 219 and 68 other proteins, respectively). On its own, this is rather weak and would only yield pfc = 0.35 to prove a physical interaction (FBS = 6.3). However, combined with other evidence such as coexpression in human and mouse, the score was substantially strengthened (FBS = 11.2; pfc = 0.98). Note that the website can also display the coupling in terms of the individual evidence or species contributions. (MEX) mRNA expression; (SLC) subcellular localization; (PPI) known protein–protein interaction; (PLC) Pearson's linear correlation coefficient; (WMI) weighted mutual information score; and PPI score. See Supplemental Methods.