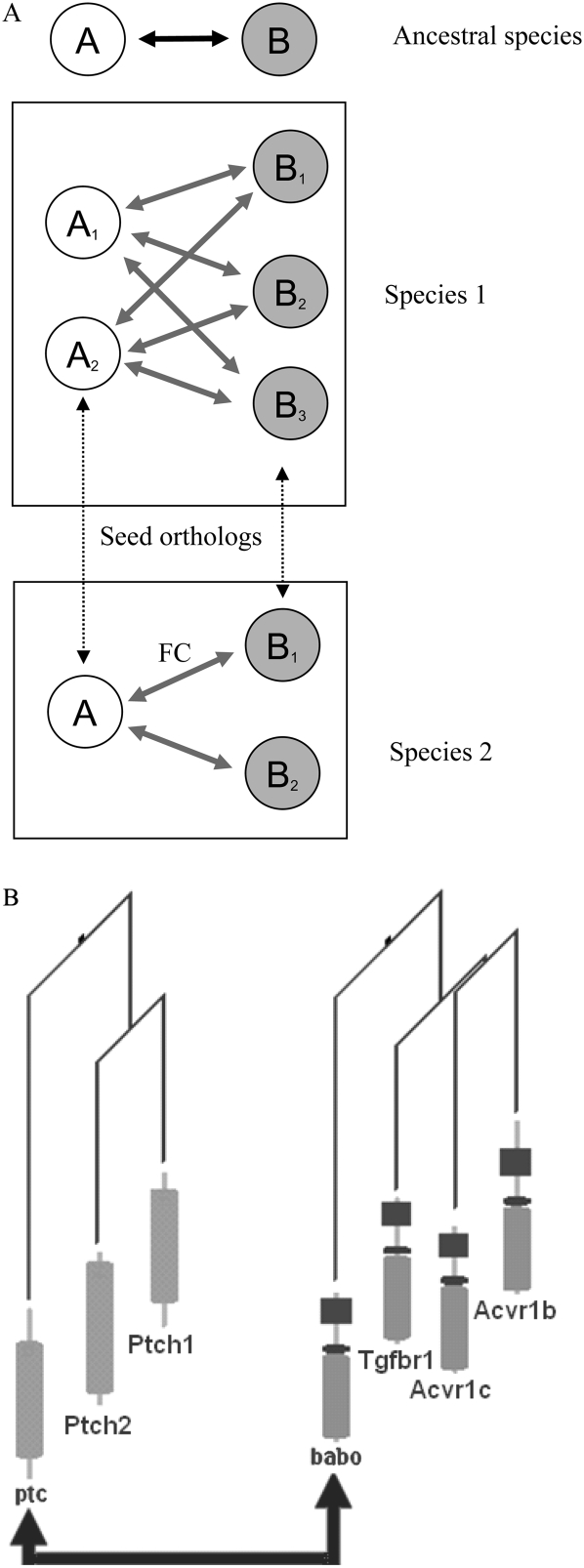
Figure 4.
Scenarios for interaction inheritance. (A) Interacting genes A and B in an ancestral species are duplicated into two and three genes in species 1, while B is duplicated into two genes in species 2. These duplicated genes are clusters of inparalogs in relation to the other species, meaning that they are co-orthologous to the corresponding A and B genes in that species. When transferring functional coupling (arrow marked FC) between orthologs, one can consider the interaction either to be valid for all inparalogs in a cluster, or to be specific for a particular inparalog pair (e.g., the seed orthologs, i.e., the two most similar ones, dotted arrows). The transfer of interaction information thus can either be done from an arbitrary inparalog in a cluster, which maximizes the coverage, or only be transferred between, e.g., seed orthologs. Benchmarking showed that transferring the best interaction in a set of inparalogs to all inparalogs in the other species yields the best results (Supplemental Material). (B) Real example of this situation: An interaction in fly ptc–babo (Shyamala and Bhat 2002) can either be considered to be valid for all six pairs of mouse inparalogs {Ptch1, Ptch2} vs. {Tgfbr1, Acvr1b, Acvr1c} or to be specific for a particular inparalog pair. Seed orthologs in this case are ptc/Ptch1 and babo/Tgfbr1. The two fly genes are connected with solid arrows; the others are mouse genes. The arrow denotes the known protein–protein interaction between ptc and babo. The leaves of the gene trees present protein domain architectures.