Figure 1. FKBP models closely represent previously determined crystal structures.
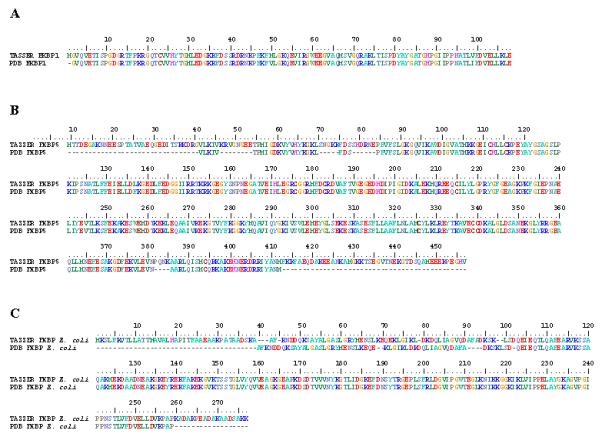
Pairwise structural alignments using ClustalW_3D between empirically generated protein structures and in silico derived models revealed that the TASSER set of programs creates complete protein representations that match closely the known crystal structures. TASSER models are on the top, with the PDB structures at the bottom of each pair. The number bar at the top indicates position of amino acids. A. TASSER generated model of FKBP1 from H. sapiens matches exactly to that of its corresponding conformation in the PDB (PDB ID: 1FKK), with the exception of a single Met in the PDB structure. B. Both N- and C-terminal regions of FKBP5 from H. sapiens are missing in the crystallograph (dashes) compared to the TASSER model. The structural alignment delineates 4 additional internal regions that are incomplete in the PDB conformation. C. Although the N- and C-termini of the E. coli FKBP from TASSER is more complete, the first 102 amino acids are shifted by 1 to 4 positions compared to the PDB structure.
