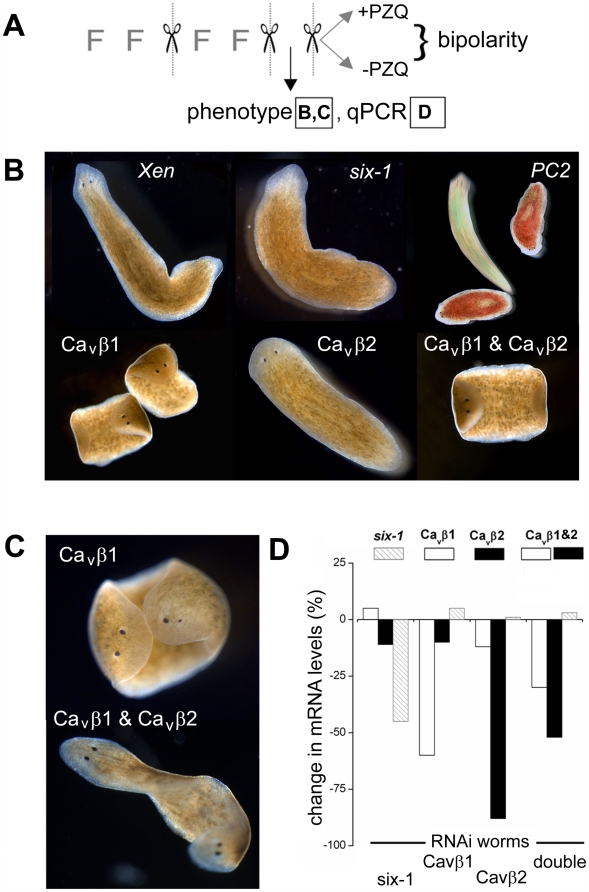
Figure 4. RNAi of Cavβ subunits in D. japonica.
(A) Schematic overview of protocol for in vivo RNAi assays. Phenotype of worms was scored after two cycles of feeding (≤2 feedings) and cutting. At this point, a sample of worms (≤10 worms) were used for qPCR, and the remaining worms were split equally into two cohorts to assay regeneration in the presence or absence of PZQ. Regenerative phenotypes are shown in (B & C) and qPCR data in (D). (B) Images of RNAi phenotypes using indicated constructs after two regenerative cycles. For the PC2 panel, worms were stained with food coloring to identify control (green) and PC2 RNAi-treated cohorts (red) and a snapshot taken over a long exposure to highlight the difference in mobility between these groups. (C) Bipolar planarian produced via Cavβ1 (top), or dual Cavβ1 and Cavβ2 knockdown (bottom) in the absence of PZQ exposure. (D) qPCR of abundance of indicated mRNAs(legend at top) in worms fed indicated dsRNA constructs (bottom). ‘Double’ represents co-feeding with Cavβ1 and Cavβ2.